Lipase variants for pharmaceutical use
A lipase and drug technology, applied in the field of determination, can solve problems such as undescribed drug use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment approach
[1181] (a) purify the lipase used in step ii); (b) dilute the lipase locally, usually 25 to 200 times in a diluent (for example 0.01% Triton-X100, 10 mM NaCl), for example to about 8 μg / mL; (c) using an extinction coefficient of 1.24A 280 / mg The concentration of the purified lipase sample was determined from the absorbance at 280nm; (d) an OD of about 0.100-0.475 in the linear range; and / or (e) the bile salt was Sigma B-8756, made 20 mM in distilled water.
[1182]In a further particular embodiment (f) of step vi), the proportion of the activity in the presence of bile salts at pH 5.0 is expressed as a percentage by calculating step v corrected for time and dilution of the "bile salt" activity ) divided by the mean of all linear data corrected for time and dilution of "no bile salt" activity.
[1183] (A) In a first specific embodiment, the lipase of the invention is selected from lipases having the following substitutions, preferably a set of substitutions, as compared to t...
Embodiment 1
[2437] Example 1: Enzyme Assay
[2438]Methods for the determination of lipase, protease and amylase activity in porcine pancreatin have been published by FIP (Fédération Internationale Pharmaceutique) as well as European Pharmacopoeia and United States Pharmacopoeia. Such assays are described eg in: Fédération Internationale Pharmaceutique, Scientific Section: International Commission on Standardization of Enzymes for Pharmaceutical Use. a) "Pharmaceutical Enzymes," Editors: R. Ruyssen and A. Lauwers, E. Story Scientia, Ghent, Belgium (1978), b) European Pharmacopoeia. See also Deemester et al. in Lauwers A, Scharpé S (eds): Pharmaceutical Enzymes, New York, Marcel Dekker, 1997, pp. 343-385. Suitable enzyme standards can be obtained from the International Commission on Pharmaceutical Enzymes, Centre for Standards, Harelbekestraat 72, B-9000 Ghent.
[2439] The lipase FIP assay and other suitable assays for lipases, proteases and amylases are described below.
[2440] Lip...
Embodiment 2
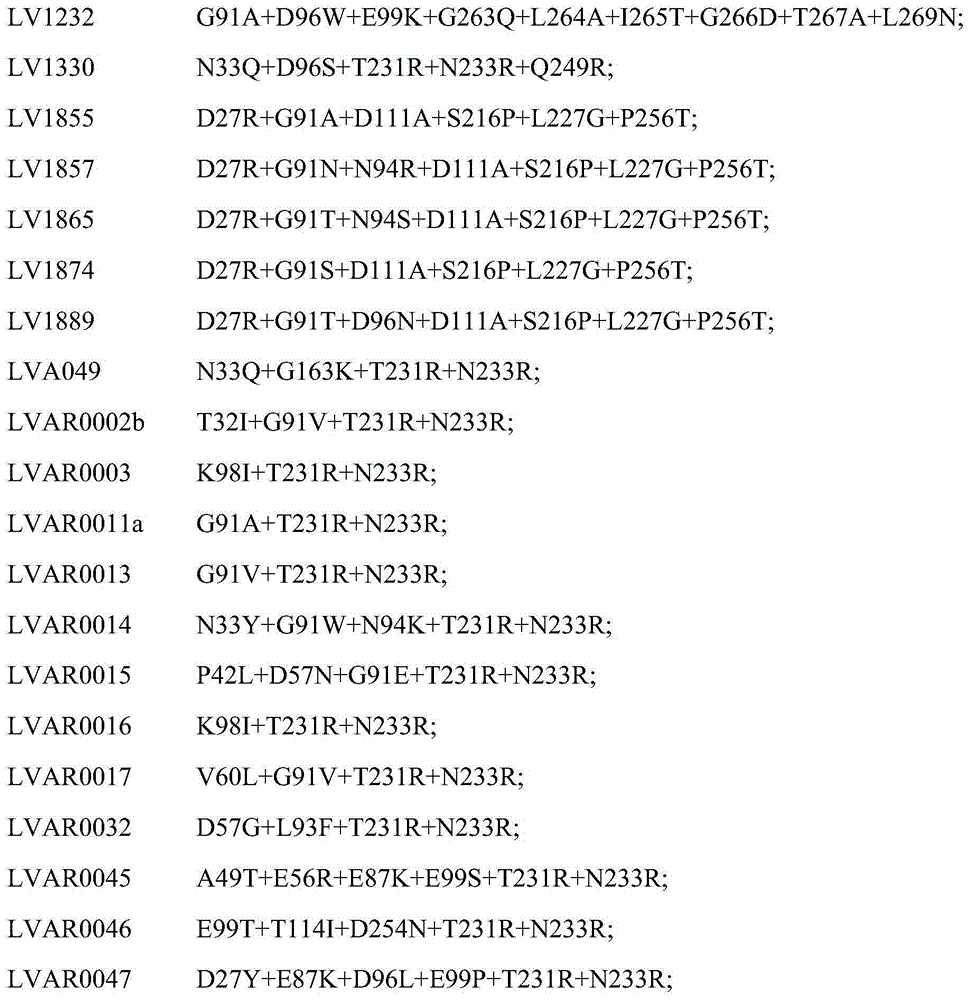
[2469] Example 2: Lipase variants with improved phospholipase activity
[2470]DNA encoding the lipase variants shown in Table 1 below was transformed into Aspergillus oryzae strain ToC1512 (described in WO2005 / 070962) using the method described in Example 22 of U.S. Patent No. 5,869,438 except that PyrG selection (described in WO2004 / 069872) instead of AMDS selection. Spores of the Aspergillus oryzae host were obtained from an agar slant and used to inoculate 10ml of YPM (10g yeast extract, Difco + 20g peptone, Difco, add water to 1 L, autoclave it; add sterile-filtered maltose to 2% (w / w )). The inoculated tubes were incubated in a New Brunswick Scientific Innova 2300 shaker at 180 rpm at 30°C for 3 days. The supernatant was harvested by filtration of the culture through Mira-Cloth (Calbiochem) followed by sterile filtration through a 0.45um (micrometer) filter. The lipase variants were purified as generally described in Example 23 of US Patent No. 5,869,438.
[2471] ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com