Integrin binding peptide and use thereof
A technology for integrating proteins and binding peptides, which can be used in the field of protein therapy and can solve problems such as ineffectiveness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0083] Production of Human Recombinant Integrin α2I Region
[0084] DNA encoding the α2I region was prepared by PCR using human integrin α2 cDNA as a template. (Integrin α2 cDNA was a gift from Dr. M. Hemler, Dana Farber, Boston). The forward primer is 5'-CACAGGGATCCCCTGATTTTCAGCTC-3' (SEQ ID No. 9), and the reverse primer is 5'-GTGGCTGAATTCAACAGTACCTTCAATG-3' (SEQ ID No. 10). Primers were designed to introduce two restriction sites in the product: a BamHI-site at the 5' end and an EcoRI-site at the 3' end. The PCR product and pGEX2T (Pharmacia) were digested with BamHI and EcoRI, ligated and transformed into E. coli DHα5F' cells followed by sequencing. The plasmid (pJKα2I) with the α2I region insert was transformed into E. coli BL2I to produce the recombinant protein rα2I. Preparation and purification of glutathione S-transferase-rα2I fusion protein was performed as follows: typically 400 ml LB (carbenicillin 50 μg / ml) was inoculated with 40 ml of an overnight culture of B...
Embodiment 2
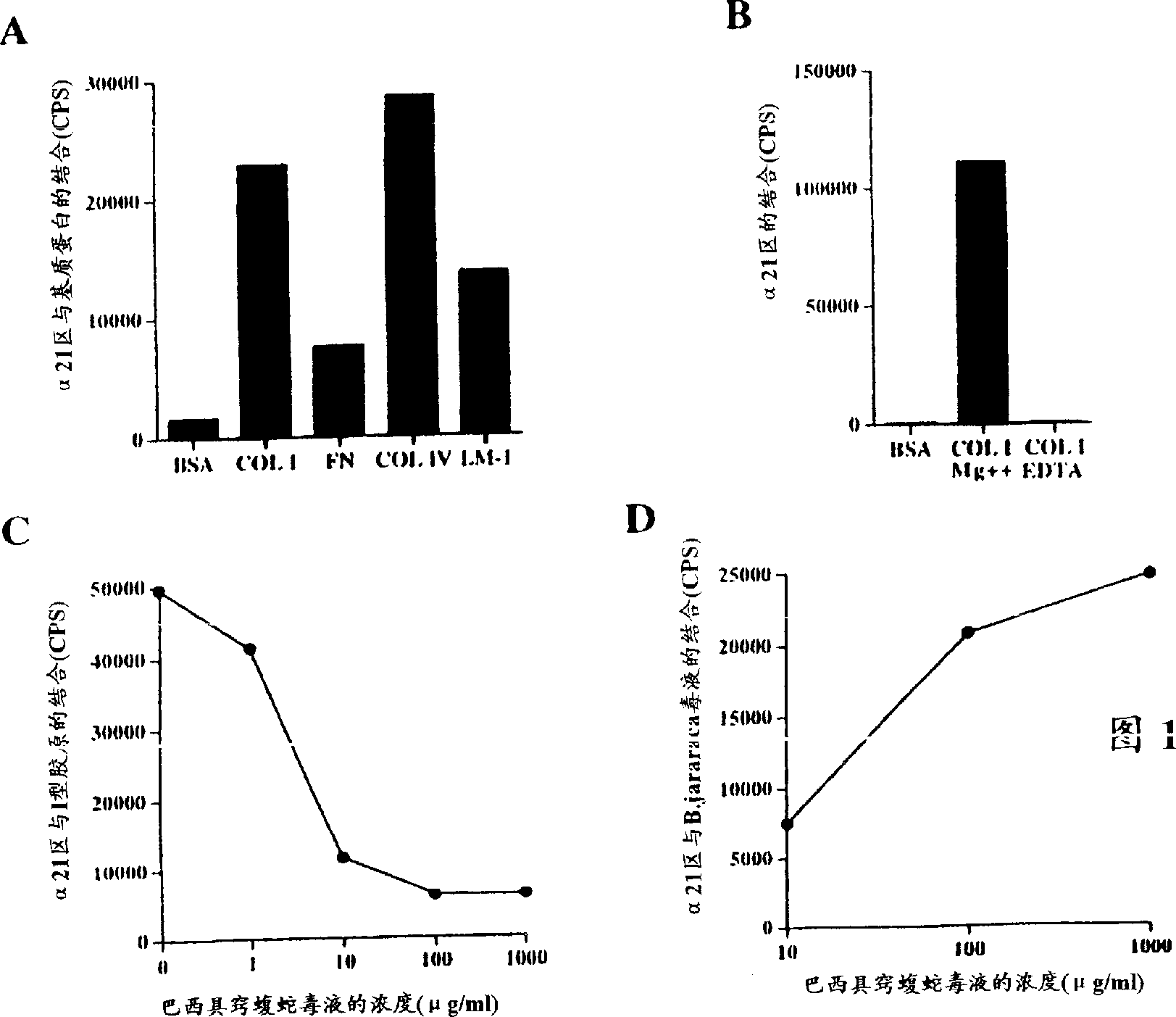
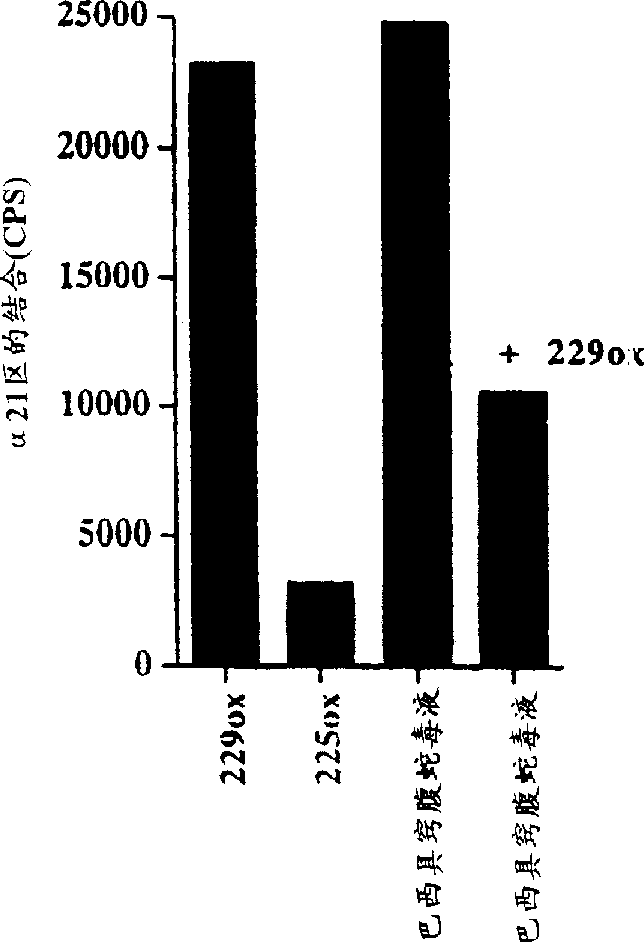
[0089] Binding assay of europium-labeled rα2I
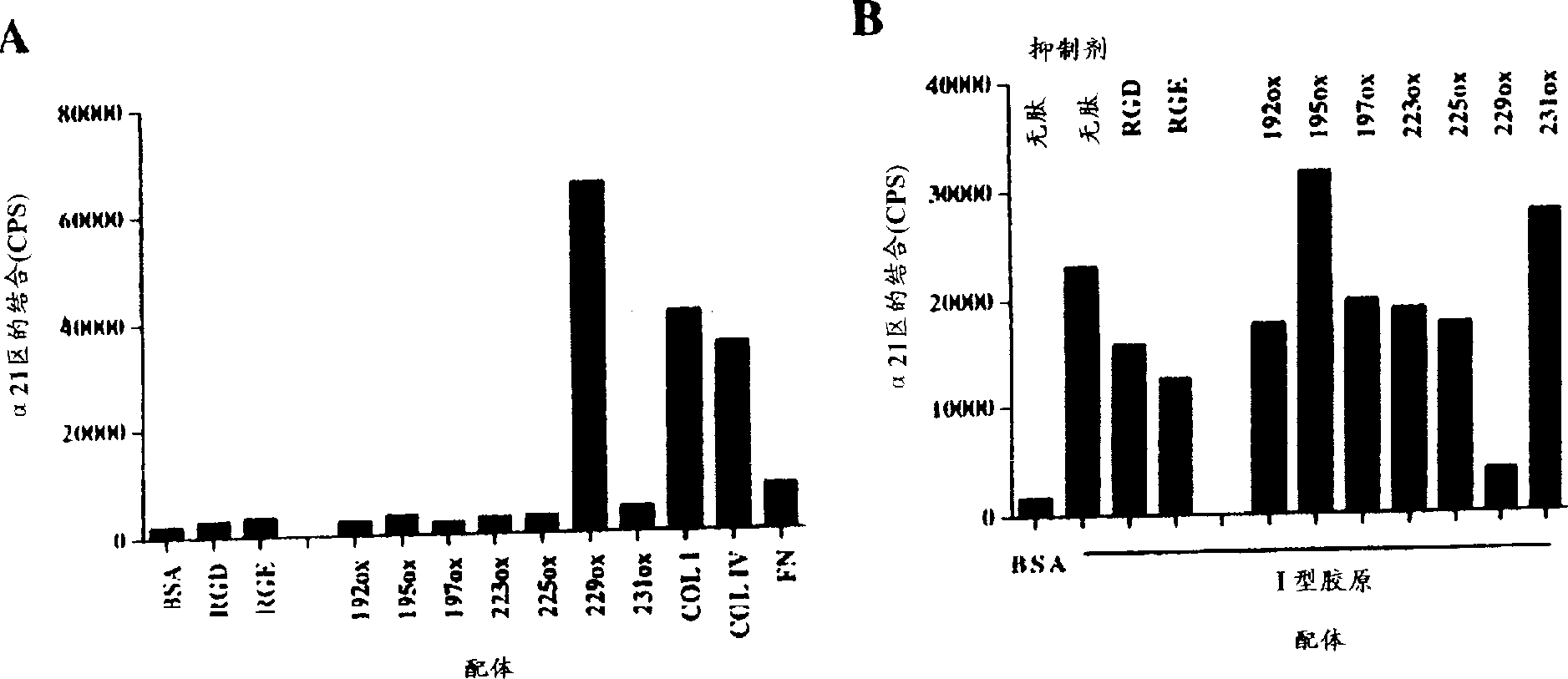
[0090] Venom from Agkistrodon brasiliensis prevents binding of recombinant integrin α2 domain I to collagen type I
[0091] A sensitive solid-phase rα2I ligand binding assay based on the use of europium-labeled rα2I was developed. Europium labeling of rα2I was performed as follows: 1 / 20 volume of 1M NaHCO was added to purified rα2I 3 (pH 8.5) to raise the pH for labeling with isothiocyanates. Europium-labeled reagent (Wallac) was added in a molar excess of 100-fold and incubated overnight at 4°C. Unbound markers were removed by gel filtration through a Sepharose G50 / Sepharose 6B (Pharmacia) column, and fractions containing marker proteins were collected.
[0092] Cover a 96-well immunoplate (Maxisorp, Nunc) by exposing the surface of each well to 0.1 ml PBS containing 5 μg / cm 2 Type I collagen on the surface of the plate. (Cowhide, Cellon), type IV collagen (Sigma), laminin-1 (purified from Engelbreth-Holm-Swarm mouse tumor ...
Embodiment 3
[0099] Peptide and binding assays using biotinylated 229ox
[0100]A Short Cyclic Jarahagin-Derived Peptide Mimics the Integrin rα2 Domain I-Collagen Interaction by Agkistrodon venom
[0101] Identification of the binding site for the α2I domain in jarahagin was performed by using a series of short cyclic peptides corresponding to the protein domain. The detection regions were selected according to the following: i) Integrin binding motifs in matrix proteins and snake venom disintegrins were found in loop structures. ii) Integrin binding motifs are known to include aspartic acid residues. iii) Published integrin-collagen interaction models also emphasize the role of arginine residues in addition to aspartate residues.
[0102] Jarahagin-derived peptides are designed based on the secondary structure of the jarahagin amino acid sequence. Secondary structure was predicted using the peptide structure program from the Genetic Computer Generating Group (GCG) software package (Mad...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com