Restrictive amplification method
A restriction and restriction enzyme digestion technology, applied in the field of restriction amplification, can solve the problems that hinder the rapid promotion of DNA microarray chip technology, and achieve the effects of accelerating research and industrialization process, uniform length, and improving signal-to-noise ratio
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
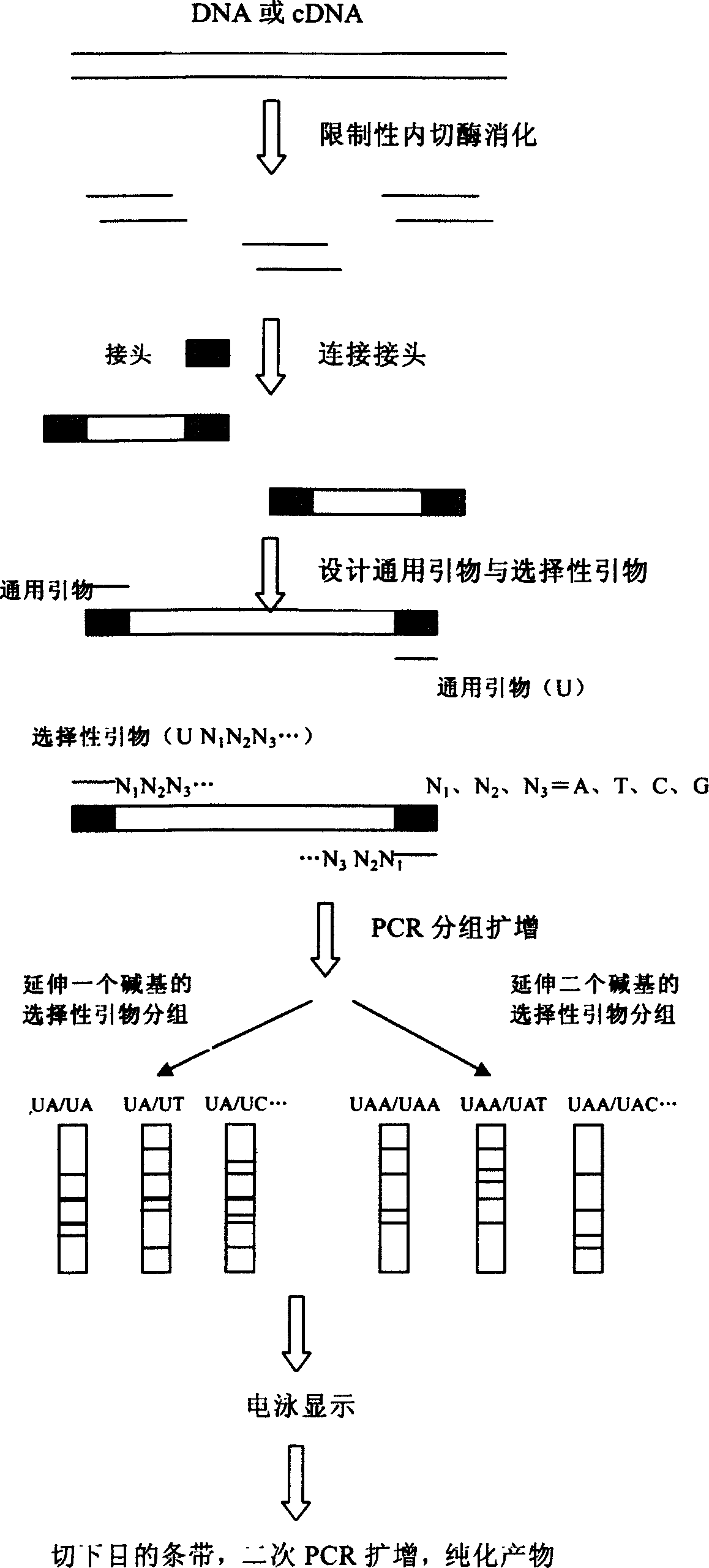
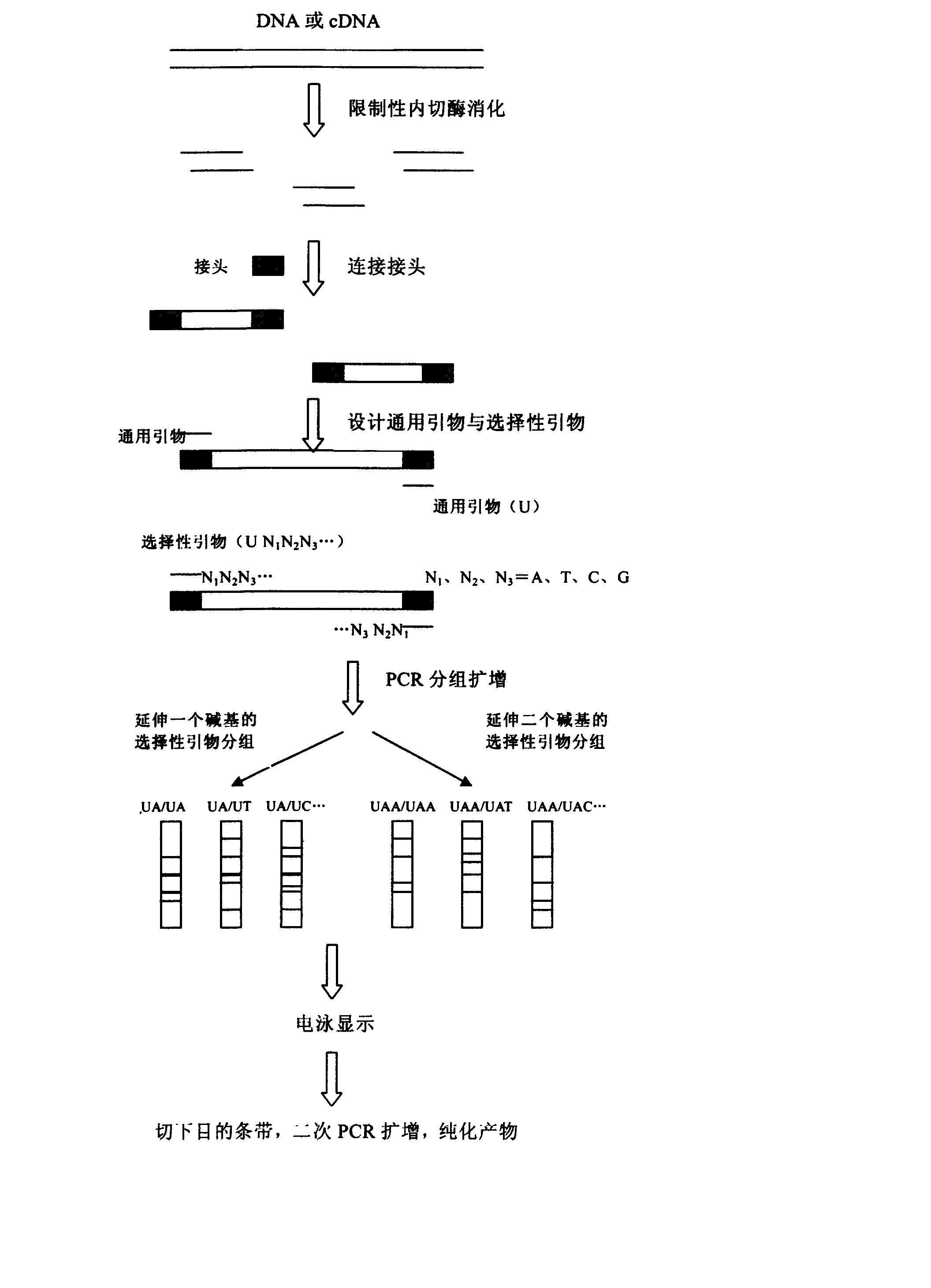
[0020] HIV1U26942 DNA is about 9000 bp in length. Analysis of its sequence shows that there are 24 restriction endonuclease Sau3A I recognition sites on the gene. 25 restriction fragments are generated after cutting with this enzyme, of which more than 100 base pairs There are 19 fragments. Analyze the sequence of the first base after GATC at both ends of the digested fragment, and perform PCR reaction with selective primers that extend by 1 base. After electrophoresis recovery, the restriction gene fragment can be quickly prepared. Specific steps are as follows:
[0021] (1) Cut the DNA with restriction endonuclease Sau3A I to generate restriction fragments with "GATC" sticky ends. The enzyme digestion reaction can refer to the following method: take about 1μg HIV1U26942DNA, add 2μl 10× endonuclease buffer, 1μl Sau3A I, add double distilled water (ddH 2 O) To 20μl, react at 37°C for 1 to 2 hours.
[0022] (2) Design two oligonucleotide chains S2P (5′-GAT CCC CCC CCCCCC CCC CCA-3′...
Embodiment 2
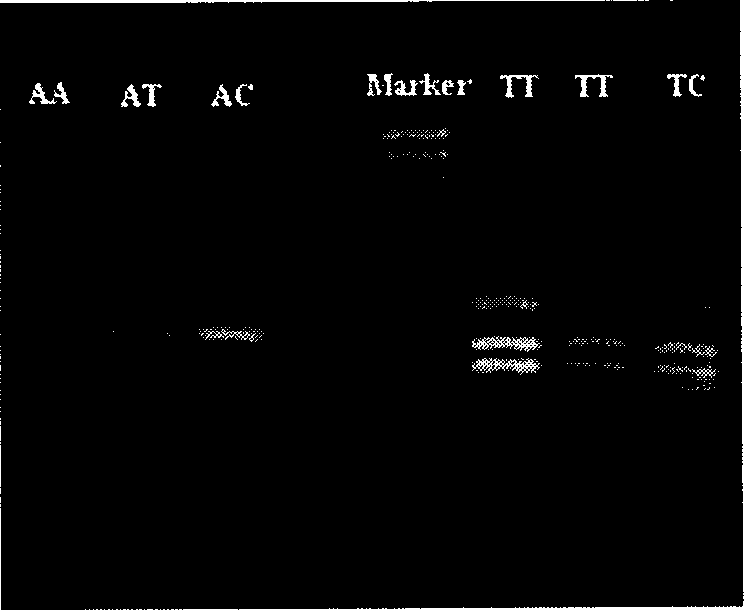
[0049] The difference between this example and example 1 is: in step (3), the two ends of the restriction fragment generated by Sau3A I digestion of HIV1U26942 DNA are analyzed for the first two bases after GATC, as shown in example 1. , The fragment was amplified by PCR with UAA and UCC, which was extended by two bases. Therefore, the above 19 fragments can be amplified by 16 PCR reactions with 13 primers that extend two bases, of which 13 reactions amplify a single band, and the other 3 PCR reactions amplify a double band ( TT1610 and TT580, TT553 and TT424, AC456 and AC441). The reaction conditions are the same as above.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com