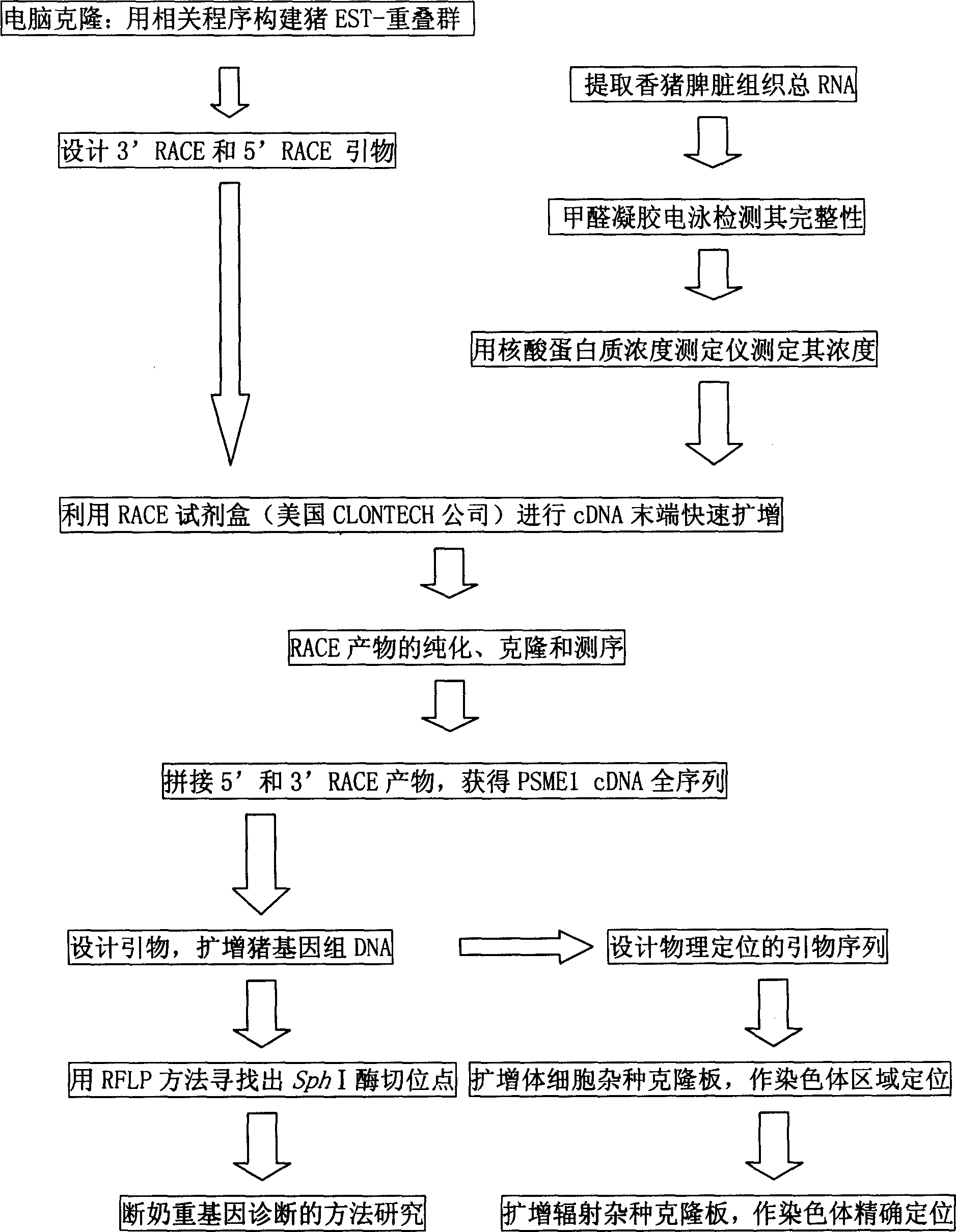
Heavy PSME1 gene for pig to pass breast, and its prepn. method
A gene and genome technology, applied in the field of porcine weaning weight PSME1 gene and its preparation, can solve problems such as research blanks
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
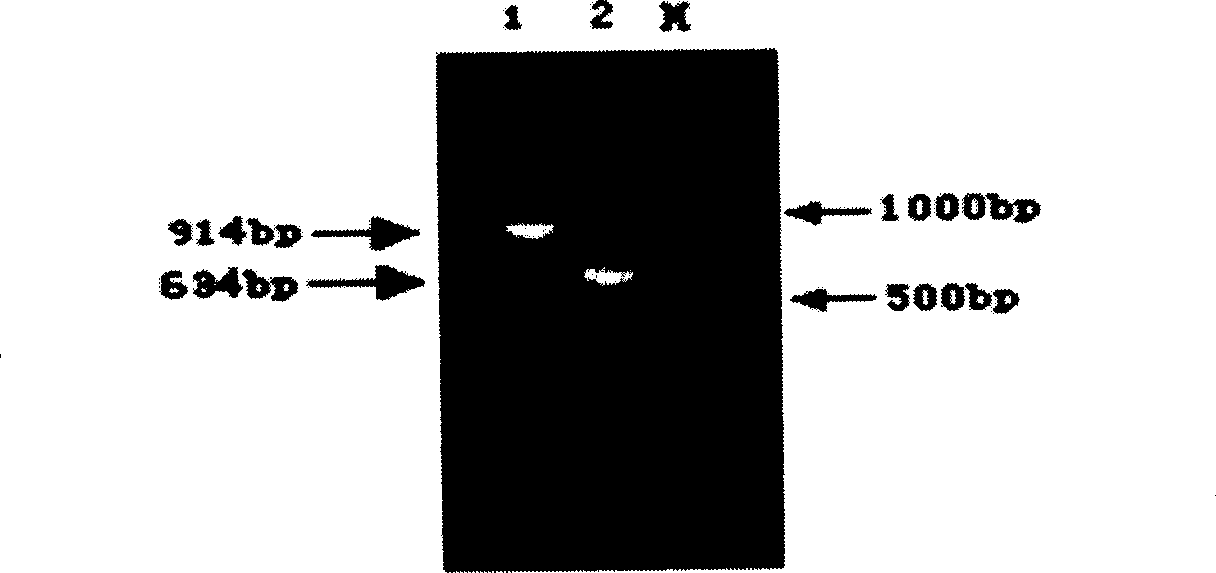
[0116] The distribution of PCR-RFLP-Sph I polymorphism in three pig breeds
[0117] (1) Primer sequence: 5'-TGAAAAGAAGAAGGGGGAAG-3' (forward, as shown in the sequence table SEQ ID: 3),
[0118] 5'-GTAAGCATTGCGGATCTCCA-3' (reverse, as shown in the sequence listing SEQ ID: 4)
[0119] (2) PCR amplification conditions: The total volume of PCR reaction is 20μl, including about 100ng of porcine genomic DNA, containing 1×buffer (Promega), 1.5mmol / L MgCl 2 , the final concentration of dNTP is 150 μmol / L, the final concentration of primer is 0.2 μmol / L, and 2U Taq DNA polymerase. Amplification program: 94°C for 4min, then 35 cycles of 94°C for 40s, 62°C for 45s, 72°C for 1min, and finally 72°C extension for 5min. PCR reaction products were detected by 2% agarose gel electrophoresis.
[0120] (3) RFLP detection conditions: Enzyme digestion reaction volume is 15μl, including 1.5μl of 1×buffer, 3~5μl of PCR product, 0.5μl (5U) of restriction endonuclease Sph I, make up 15...
Embodiment 2
[0130] We used the established PCR-RELP diagnostic method to detect the genotype of PSME1-RFLE-SphI on 190 DNA samples from a Large White herd.
[0131] The test results showed that among 190 individuals, there were 10 individuals with AA genotype, 62 individuals with AB genotype, and 118 individuals with BB genotype. In the association analysis with weaning weight, the results of the simple mean and standard deviation analysis of the traits among the genotypes are summarized in Table 4. The results of correlation analysis showed that the weaning weights of AA and AB genotype pigs were significantly different (P<0.01), and the weaning weights of AA and BB genotype pigs, and AB and BB genotype pigs were significantly different (P<0.05).
[0132] Table 3: Association analysis of different PSME1 gene SphI-RFLP genotypes and weaning weight
[0133] Genotype Genotypes Individual number N Weaning weight Weaning weight(kg)
[0134] Psme1-SphI AA 10 11.57 ± 1.51
[013...
Embodiment 3
[0141] The relationship between the genotypes of 108 Tongcheng pigs and the weaning weight of piglets was detected by the genetic diagnosis method as described above. The relationship between the genotype of Tongcheng pigs and the weaning weight of piglets is shown in Table 5.
[0142] Table 4 The relationship between the genotype of Tongcheng pigs and the weaning weight of piglets
[0143] Genotype Number of individuals Weaning weight (kg)
[0144] Psme1-SphI AA 2 10.69 ± 1.35
[0145] BB82 9.36±1.58
[0146]AB 24 10.21±1.40
[0147] P-value BB-AB 0.032
[0148] In this example, since only 2 pigs with AA type were detected, we ignored the AA type when doing correlation analysis, and re-tested the T-test only for the weaned pigs with BB-AB type. The results showed that the weaning weights of AB and BB pigs were significantly different, which was consistent with the previous results.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com