Method for determining the presence of extension products
A product, micro-sequencing technology, applied in biochemical equipment and methods, microbial determination/inspection, fermentation, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
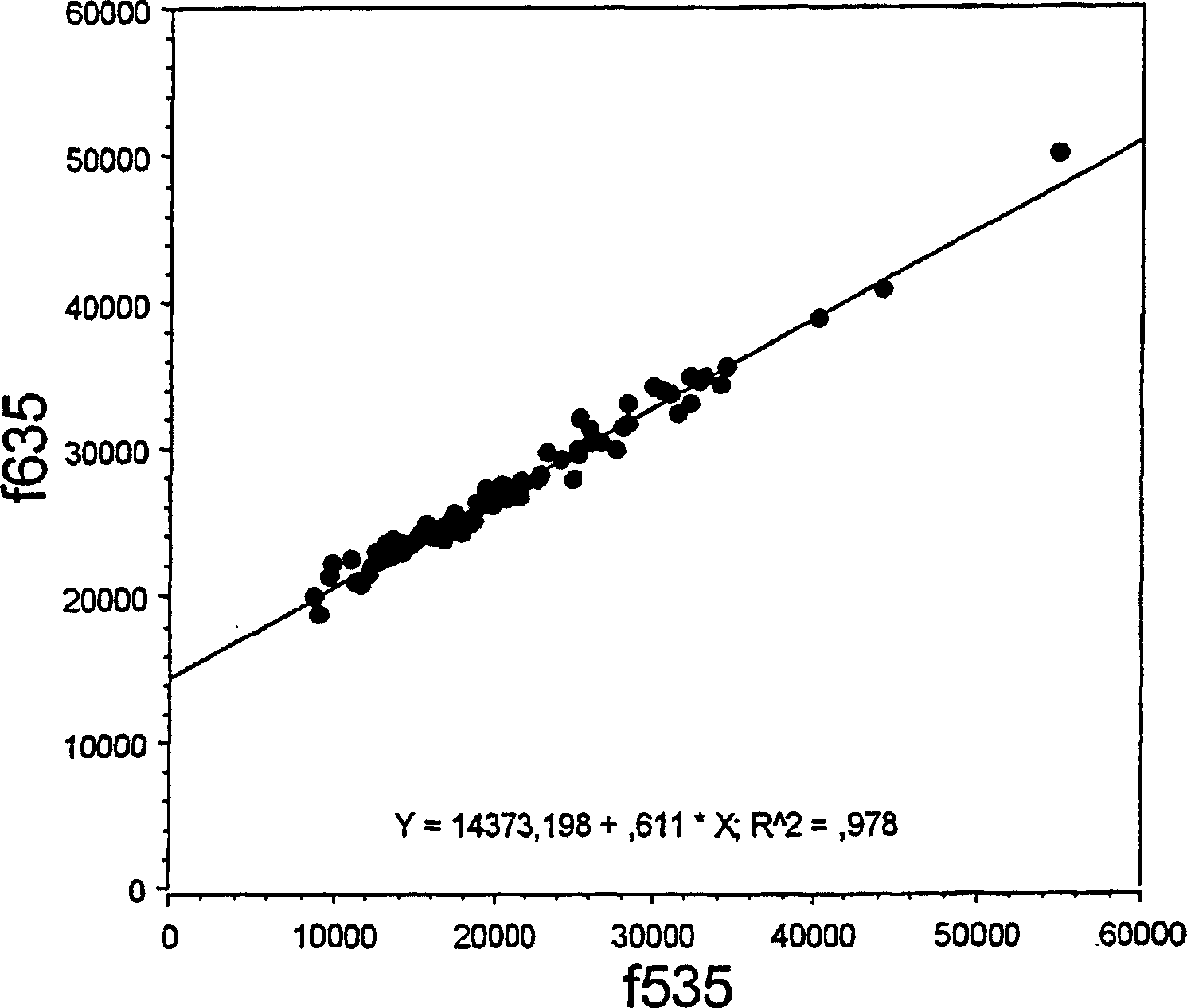
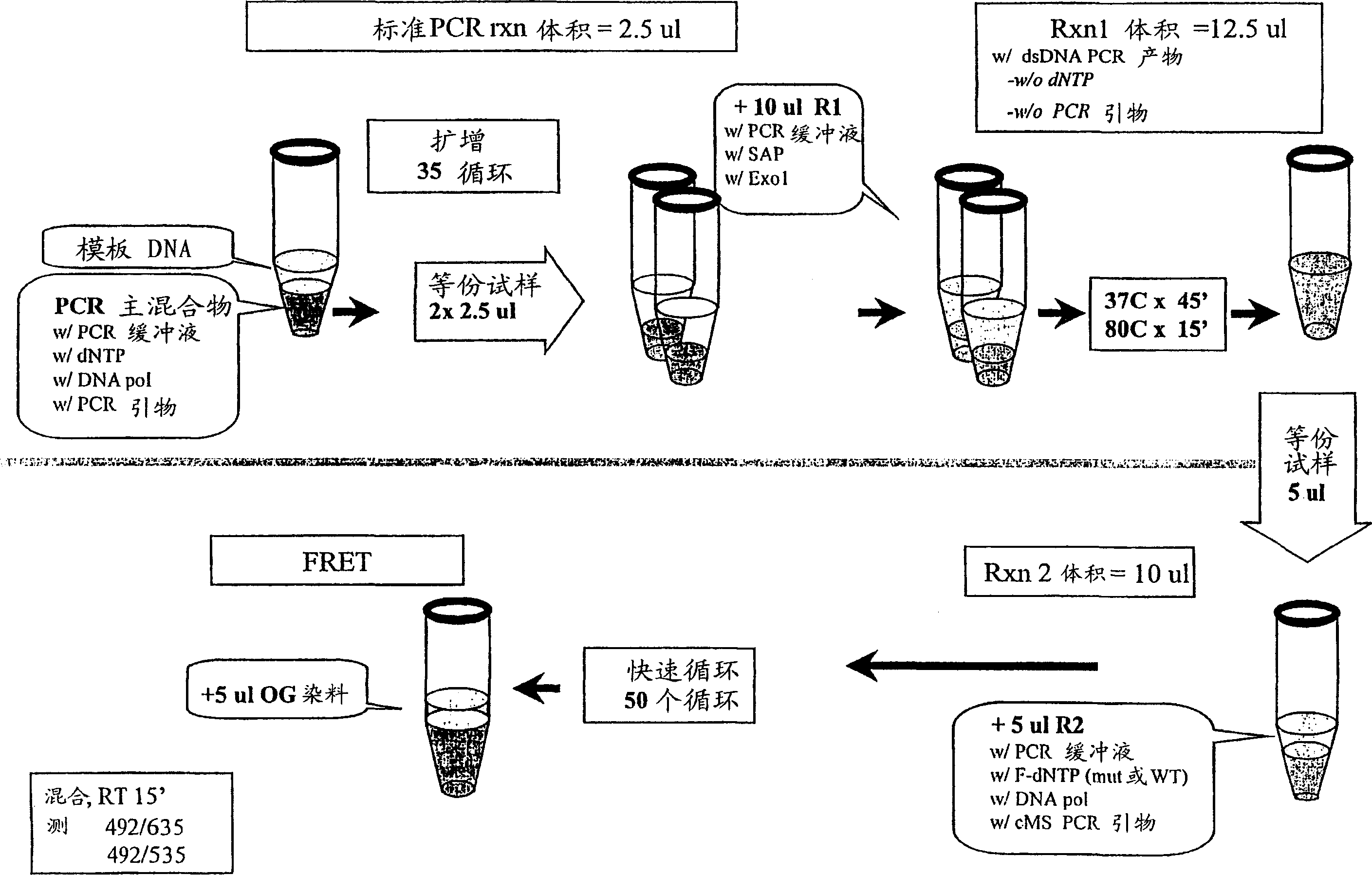
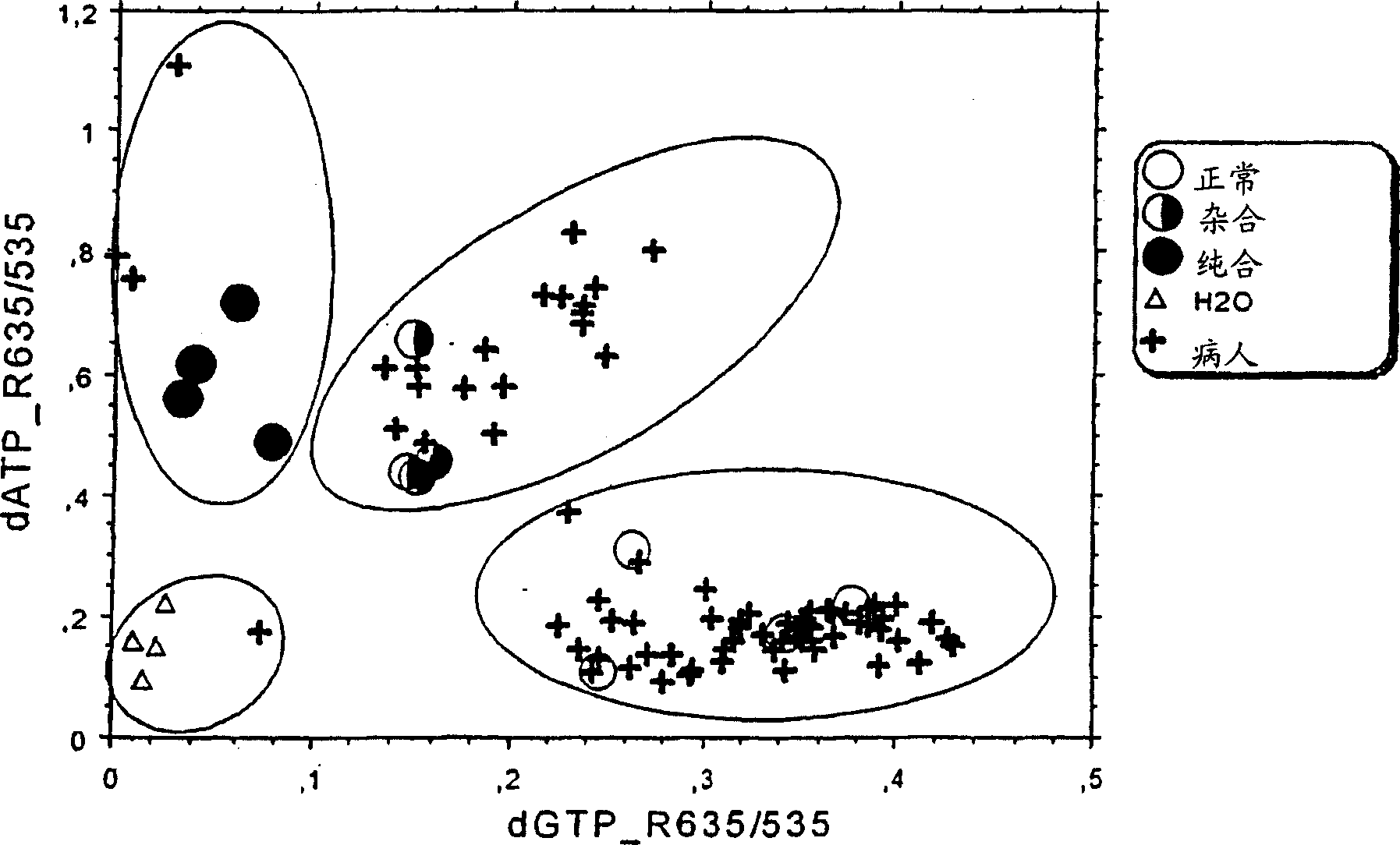
[0051] SNP genotypes were determined by cycle micro-sequencing (cMS) technology modified from the original solid-phase micro-sequencing (US Patent 6,013,431). figure 2 The procedure of Example 1 is illustrated. Template DNA was extracted from frozen EDTA blood samples by conventional methods. The primers used for PCR amplification were sequences flanking the polymorphic SNP site, which in this case was the Factor V Leiden mutation (Arg506Gln, corresponding to a nucleotide change from G to A in the coding strand of genomic DNA) , which confers protein C resistance and increases the patient's risk of trombosis. The primer sequences were 5'-TCT CTT GAA GGA AATGCC CCA-3' and 5'-GGG CTA ATA GGA CTA CTT CTA-3'. Add 5 μL of template DNA (5 ng / μL) to 40 μL of the PCR master mix previously dispensed in a 96-well PCR plate. PCR amplification was performed in DNA polymerase reaction buffer and 25 μM of PCR primers, 200 μM of dNTPs and 0.5 units of thermostable DNA polymerase were add...
Embodiment 2
[0055] DNA used as template was extracted from frozen EDTA blood samples by conventional methods. The primers used for PCR amplification were sequences flanking the polymorphic SNP site, which in this case was the Factor V Leiden mutation (Arg506Gln, corresponding to a nucleotide change from G to A in the coding strand of genomic DNA) , which confers protein C resistance and increases the patient's risk of trombosis. The primer sequences were 5'-TCT CTT GAA GGA AAT GCC CCA-3' and 5'-GGG CTA ATA GGA CTACTT CTA-3'. Add 5 μL of template DNA (5 ng / μL) to 40 μL of the PCR master mix previously dispensed in a 96-well PCR plate. PCR amplification was performed in DNA polymerase reaction buffer, and 25 μM of PCR primers, 200 μM of dNTPs and 0.5 units of thermostable DNA polymerase were added. The amplification reaction was carried out in an MJ Research Tetrad cycler with a dynamic drop PCR program at 64°C => 58°C: initially at 96°C for 5 minutes, then at 96°C for 1 minute, and withi...
Embodiment 3
[0059] We tested a variety of fluorescent dyes as acceptors to receive FRET energy from ssDNA-binding fluorescent reagent OliGreen (Molecular Probes, Eugene, OR). We took 5 μL of single-stranded DNA-specific fluorescent reagent OliGreen diluted 1:50 with TE buffer (10mM Tris-HCl, pH 7.0, 1mM EDTA), and added it to the sample containing 5pmol of primers labeled with one of the following dyes: Tetramethyl Rhodamine-based (emit around 552nm), Texas Red TM (emit around 620nm, Molecular Probe, Eugene OR), Cy3 (emit around 570nm, Amersham Lifescience, Inc.) or Cy5 TM (emission around 667nm, Amersham Life Science, Inc.). All of these dyes emit at their expected emission wavelengths when attached to oligonucleotides, and are excited at 485 nm in the presence of OliGreen bound to the primer. Thus, using ssDNA-binding fluorescent reagents and FRET, the presence of multiple oligonucleotides carrying fluorophores with different emission peaks can be detected.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com