Fluorescent quantitative PCR detecting method for hepatitis B virus and special reagent kit
A hepatitis B virus, fluorescence quantitative technology, applied in the direction of biological testing, fluorescence/phosphorescence, microbial measurement/inspection, etc., can solve the problems of not being suitable for clinical application, low specificity, etc., to shorten the extraction time and improve detection Sensitivity, simple and fast operation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Embodiment 1. Design and synthesis of hepatitis B virus fluorescent quantitative PCR primers and probes
[0038] (1) Sequence comparison: use the sequence comparison function provided by http: / / www.ncbi.nlm.nih.gov / blast website to perform HBV whole gene sequence comparison, and find out the conserved regions in the whole genome.
[0039] (2) Using Beacon Designer 2.1 (Molecular Beacon Design Software 2.1) software to design fluorescent quantitative PCR primers and probes, the designed three pairs of primers and probes are respectively located in the conserved regions of surface antigen S, core antigen C and E antigens , primers and probe sequences were synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd.
[0040] S region upstream and downstream primers and probes:
[0041] RSU: 5'AGAATCCTCACAATACCGCAGAGT 3'
[0042] RSL: 5'CACACGGTAGTTCCCCCTAGAA 3'
[0043] PS: 5'FAM-AGACTCGTGGTGGACTTCTCTCAAT-TAMARA 3'
[0044] C region upstream and downstrea...
Embodiment 2
[0052] Embodiment 2. The preparation of detection standard
[0053] (1) Extraction of viral DNA
[0054] Add 50 μl of 0.4M NaOH to 50 μl of serum from patients with hepatitis B virus, and incubate at 80° C. for 10 minutes. Centrifuge at 15,000 rpm for 30 seconds, take the supernatant, and add 25 μl of 0.4M Tris-HCl pH7.5.
[0055] (2) PCR amplification
[0056] PCR amplification was carried out with the same upstream and downstream primers as the surface antigen S, core antigen C and E antigens in fluorescent quantitative PCR.
[0057] The PCR amplification system is 25 μl, including
[0058] 10×PCR buffer 2.5μl
[0059] 2.5mM dNTP 2.0μl
[0060] Upstream primer (5μM) 1.0μl
[0061] Downstream primer (5μM) 1.0μl
[0062] Taq enzyme (5u / μl) 0.2μl
[0063] Viral DNA 2.0μl
[0064] h 2 O 16.3 μl
[0065] 25μl
[0066] 10×PCR buffer composition is 500mM / L Tris-HCl pH8.3, 500mM / LKCI, 20mM MgCI 2 . Amplification was carried out using...
Embodiment 3
[0071] Embodiment 3. Preparation of standard curve and its optimization
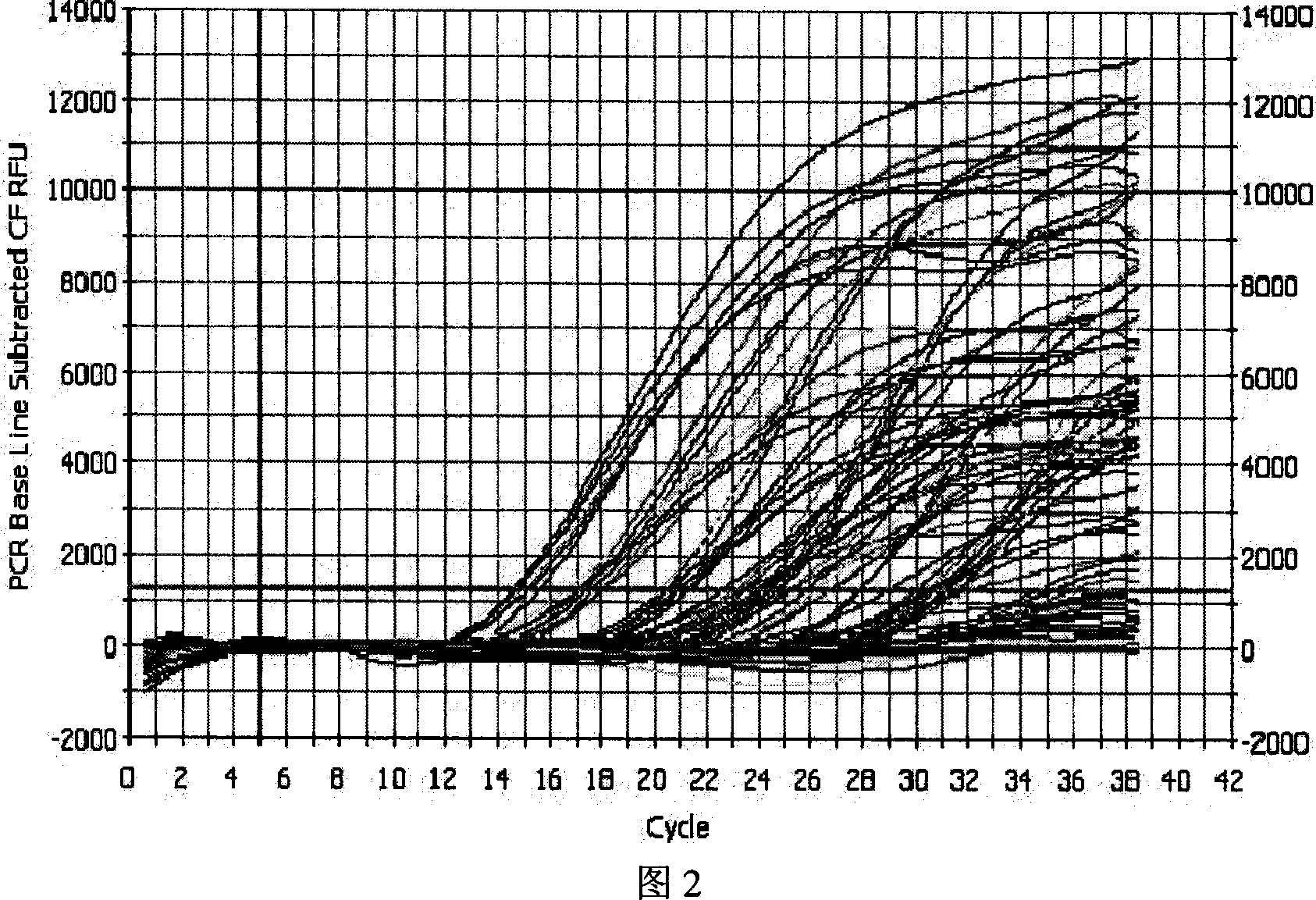
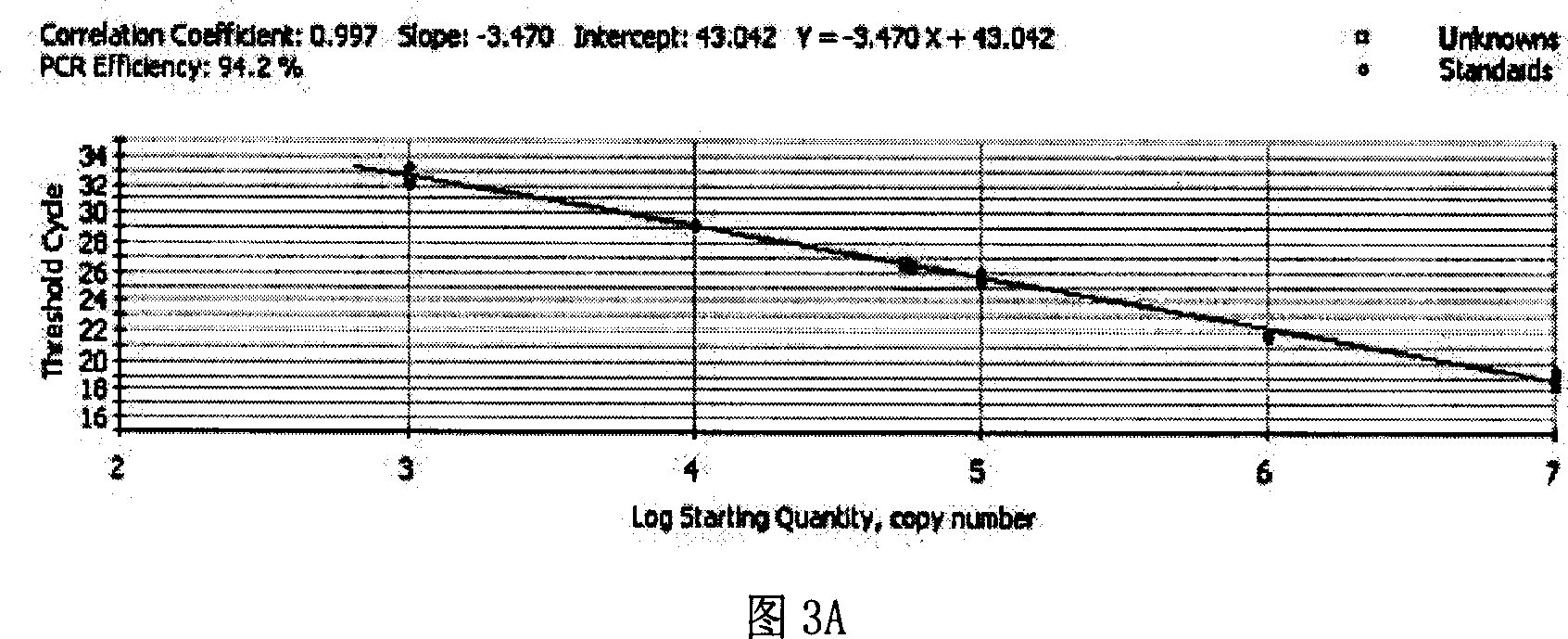
[0072] According to the copy number obtained after quantification, the samples of the three regions were made into 1×10 3 -10 7 (copies / μl) and other 5 gradiently diluted detection standards, respectively make three standard curves for three regions, and detect the Ct value of the same virus sample on three different standard curves (Ct is the cycle threshold, referring to each The number of cycles experienced when the fluorescent signal in the reaction tube reaches the set threshold value) and the copy number, so as to determine the standard curve with the best amplification effect. Among them, each sample has three repetitions. In order to reduce the sampling error and improve the repeatability of the experiment, a Thermo Electron (Saimo Company) electric sampling gun is used to add samples, and each experimental well is repeated three times.
[0073] (1) Extraction of viral DNA
[0074] Same as in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com