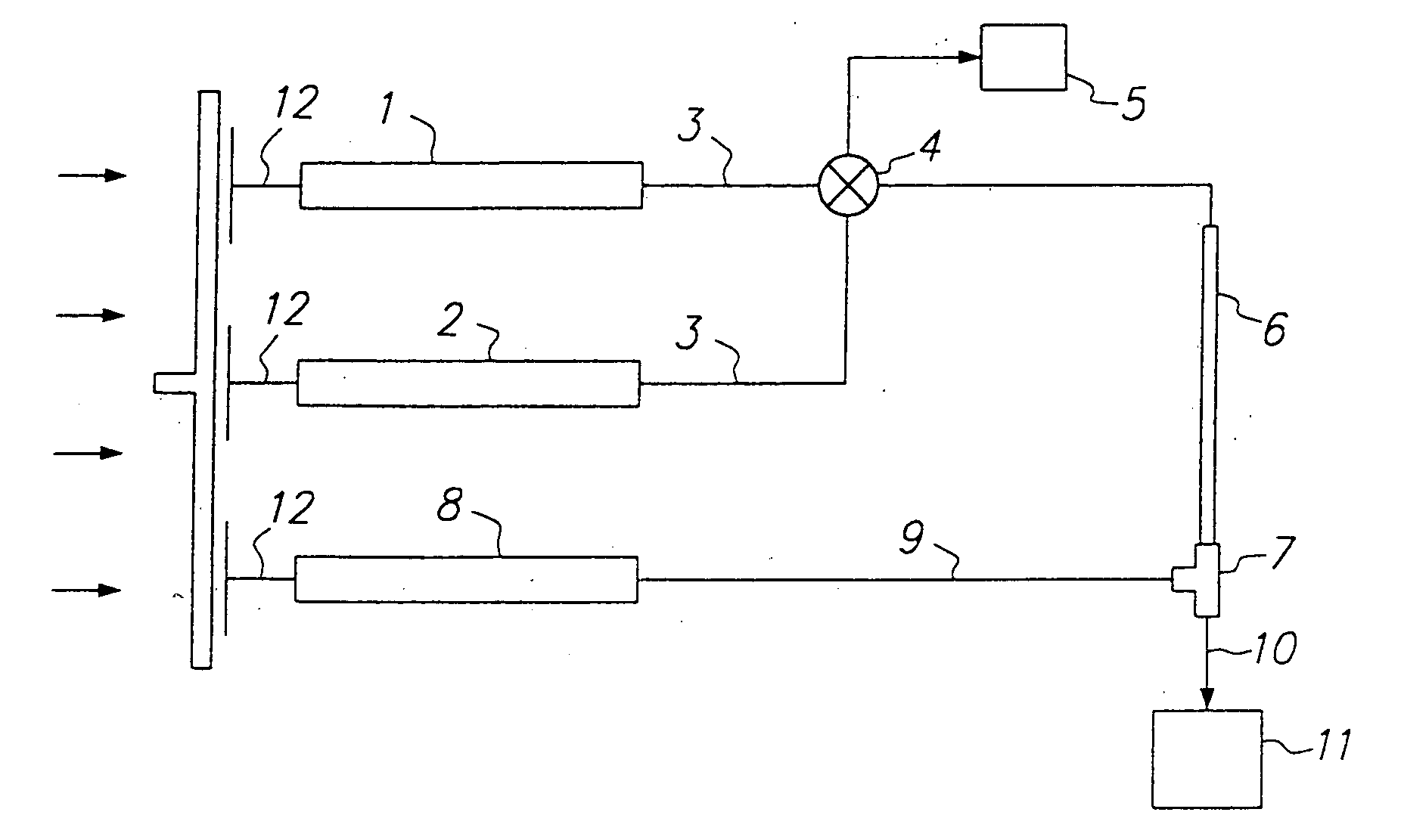
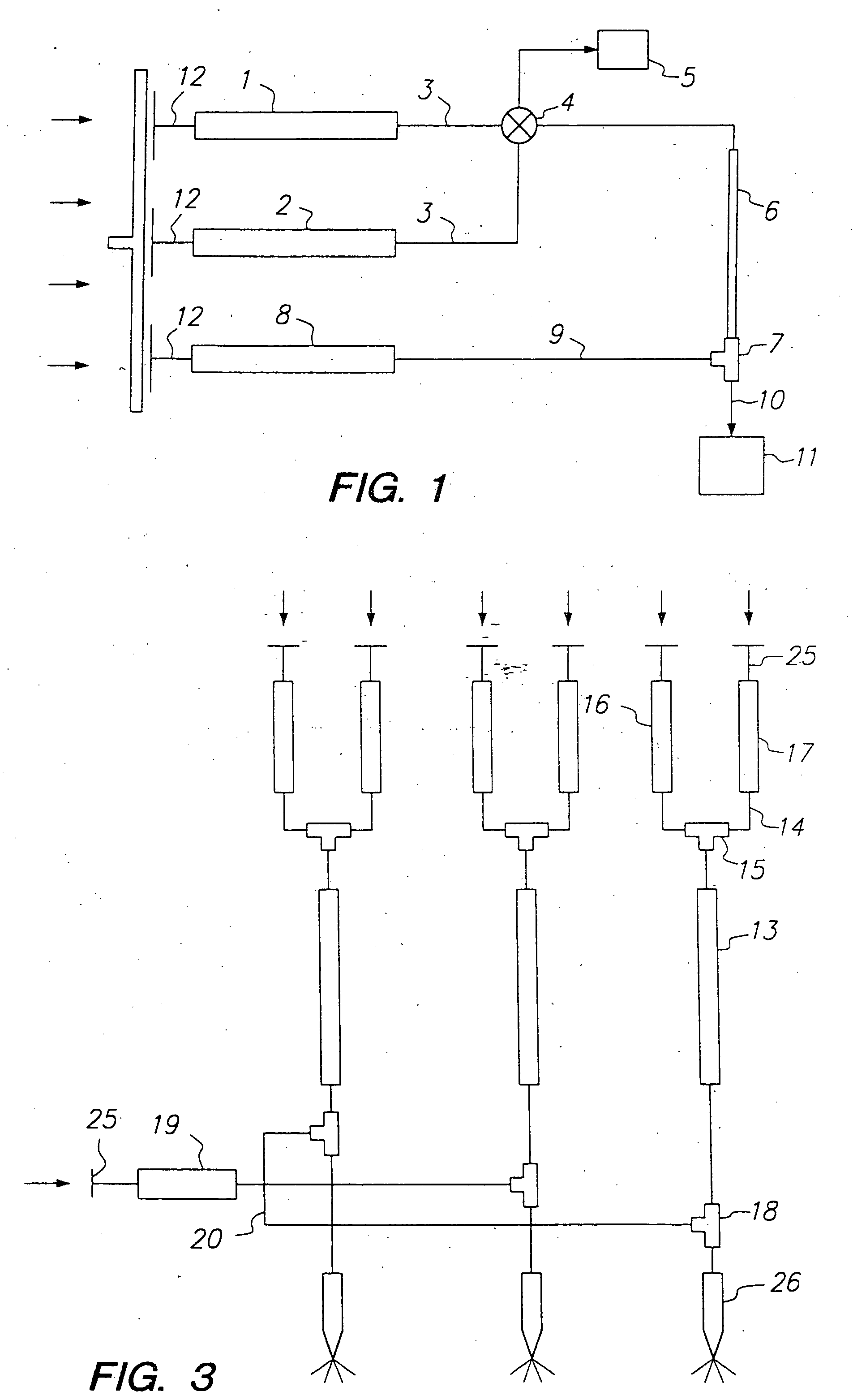
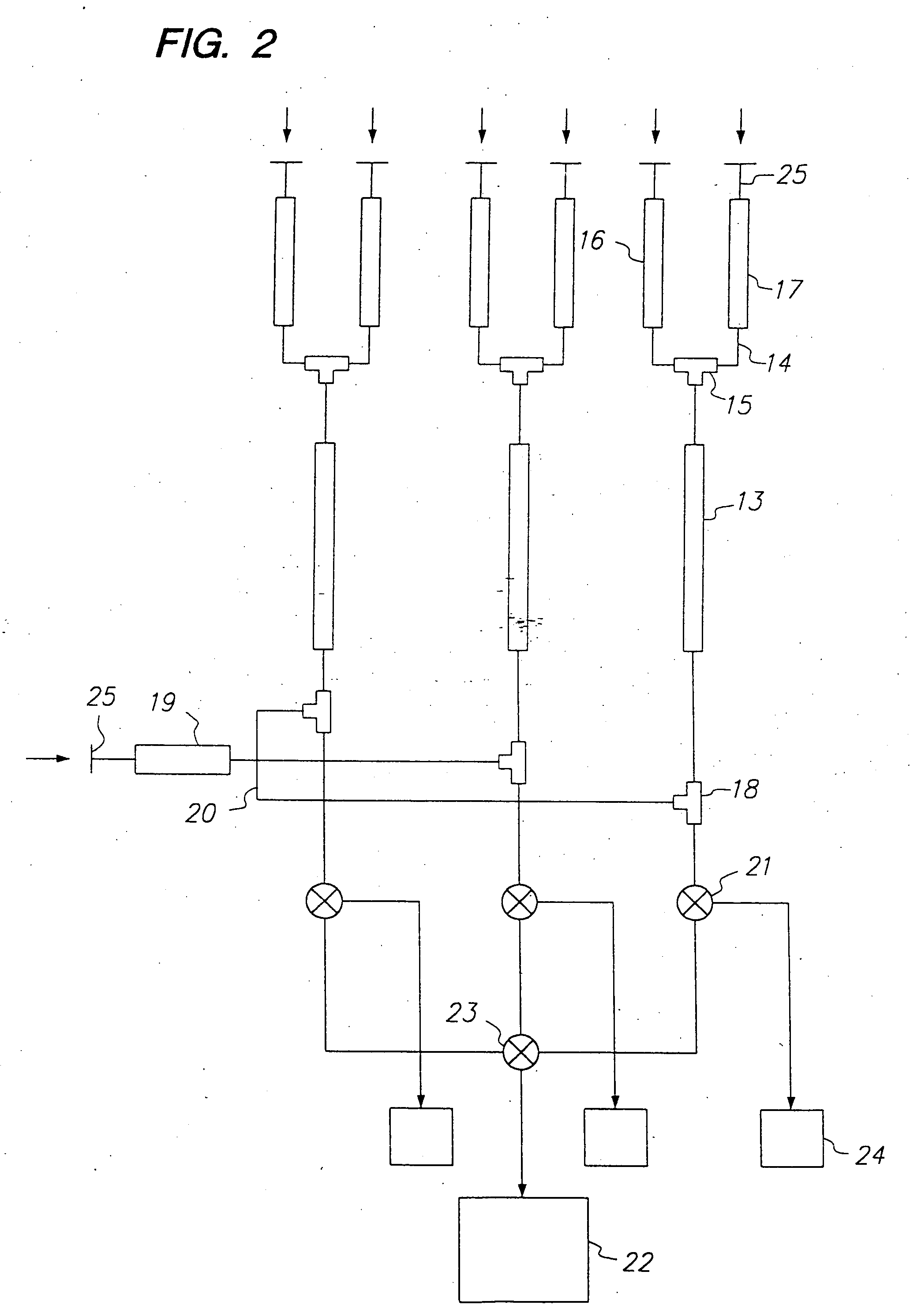
Methods for screening compound libraries
a compound library and library technology, applied in chemical libraries, combinational chemistry, component separation, etc., can solve the problems of inability to identify compounds having the desired biological activity, need to keep track, and inability to screen each compound individually, so as to reduce the screening time for each library, shorten the break through time, and achieve the effect of weak affinity for the target receptor
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Screening of an Oligosaccharide Library Using FC-MS
[0210] In this example, a compound library containing a mixture of six oligosaccharides was screened using frontal chromatography in combination with an electrospray mass spectrometer to determine the relative affinity of the oligosaccharides for a monoclonal antibody that recognizes the 3,6-dideoxy-D-galactose (abequose) epitope in Salmonella paratyphi B O-antigens.
[0211] The compound library consisted of the following six oligosaccharides: .alpha.GalNAc(1.fwdarw.3).beta.Gal-OGr (compound 1); .alpha.Gal(1.fwdarw.3)[.alpha.Fuc(1.fwdarw.2)].beta.Gal-OGr (compound 2); .alpha.Man(1.fwdarw.3)[.alpha.Man(1.fwdarw.6)].beta.Man-OGr (compound 3); .alpha.Abe(1.fwdarw.3).alpha.Tal-OCH.sub.3 (compound 4); .alpha.Gal(1.fwdarw.2)[.alpha.Abe(1.fwdarw.3)].alpha.Man-OCH.sub.3 (compound 5); and .alpha.Glc(1.fwdarw.4).beta.Glc(1.fwdarw.4).alpha.Gal(1-.fwdarw.2)-[.alpha.Abe(1.fwdarw.3)].alpha.Man(1.fwdarw.3).alpha.Glc(1.fwda-rw.4).beta.Glc-OCH.sub.3 (...
example 2
Screening of an Oligosaccharide Library Using FC-MS and an Indicator Compound
[0223] In this example, the use of an indicator compound to screen a compound library is demonstrated. The antibody used in this example was the same as that used in Example 1, i.e., a monoclonal antibody that recognizes the 3,6-dideoxy-D-galactose (abequose) epitope in Salmonella paratyphi B O-antigens. The column was also essentially the same as the column in Example 1 and it was prepared and operated as described therein.
[0224] In this experiment, three solutions were prepared. Solution A contained the following four oligosaccharide in 2 mM NH.sub.4OAc: .alpha.GalNAc(1.fwdarw.3).beta.Gal-OGr (compound 1); .alpha.Gal(1.fwdarw.3)[.alpha.Fuc(1.fwdarw.2)].beta.Gal-OGr (compound 2);
[0225] .alpha.Man(1.fwdarw.3)[(.alpha.Man(1.fwdarw.6)].beta.Man-OGr (compound 3); .alpha.Abe(1.fwdarw.3).alpha.Tal-OCH.sub.3 (compound 4), wherein Gr=O(CH.sub.2).sub.8CO.sub.2CH.sub.3. Solution B contained
[0226] .alpha.Gal(1.fwdarw...
example 3
Screening of an Oligosaccharide Library Using FC-MS
[0230] In this example, a compound library containing a mixture of four oligosaccharides was screened using frontal chromatography in combination with an electrospray mass spectrometer to determine the relative affinity of the oligosaccharides for cholera toxin B subunit.
[0231] The compound library consisted of the following four oligosaccharides: .alpha.GalNAc(1.fwdarw.3).beta.Gal-OGr (compound 1); .alpha.Gal(1.fwdarw.3)[.alpha.Fuc(1.fwdarw.2)].beta.Gal-OGr (compound 2); .alpha.Man(1.fwdarw.3)[.alpha.Man(1.fwdarw.6)].beta.Man-OGr (compound 3); and GM.sub.1 oligosaccharide (compound 7, wherein Gr=O(CH.sub.2).sub.8CO.-sub.2CH.sub.3. Compound 7, which is the natural ligand for cholera toxin B subunit, was obtained using the procedures described in A. Schon et al., "Thermodynamics of Intersubunit Interactions in Cholera Toxin upon Binding to the Oligosaccharide Portion of Its Cell Surface Receptor, Ganglioside G.sub.M1" Biochem. 1989, ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| internal diameter | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| internal diameter | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com