Method for quantitative measurement of gene expression for indentifying individuals at risk for bronchogenic carcinoma
a bronchogenic carcinoma and indentification method technology, applied in the field of quantitative measurement of gene expression for indentification individuals at risk of bronchogenic carcinoma, can solve the problems of generating data that are difficult to interpret, reducing the quantitative level of gst or glutathione peroxidase gene expression in primary, and increasing the risk of oxidative damage of normal bronchial epithelial cells (nbecs)
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
Materials and Methods
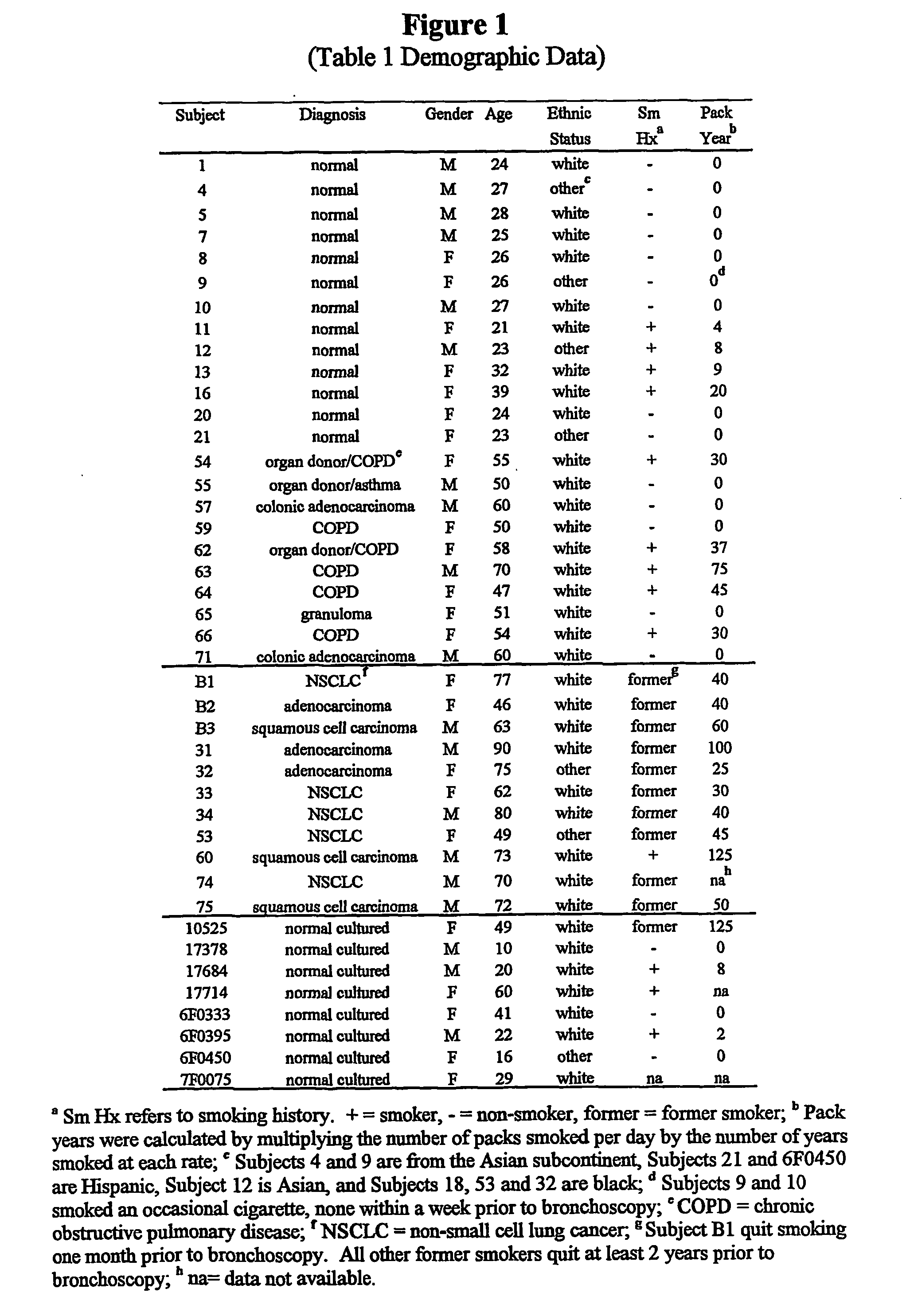
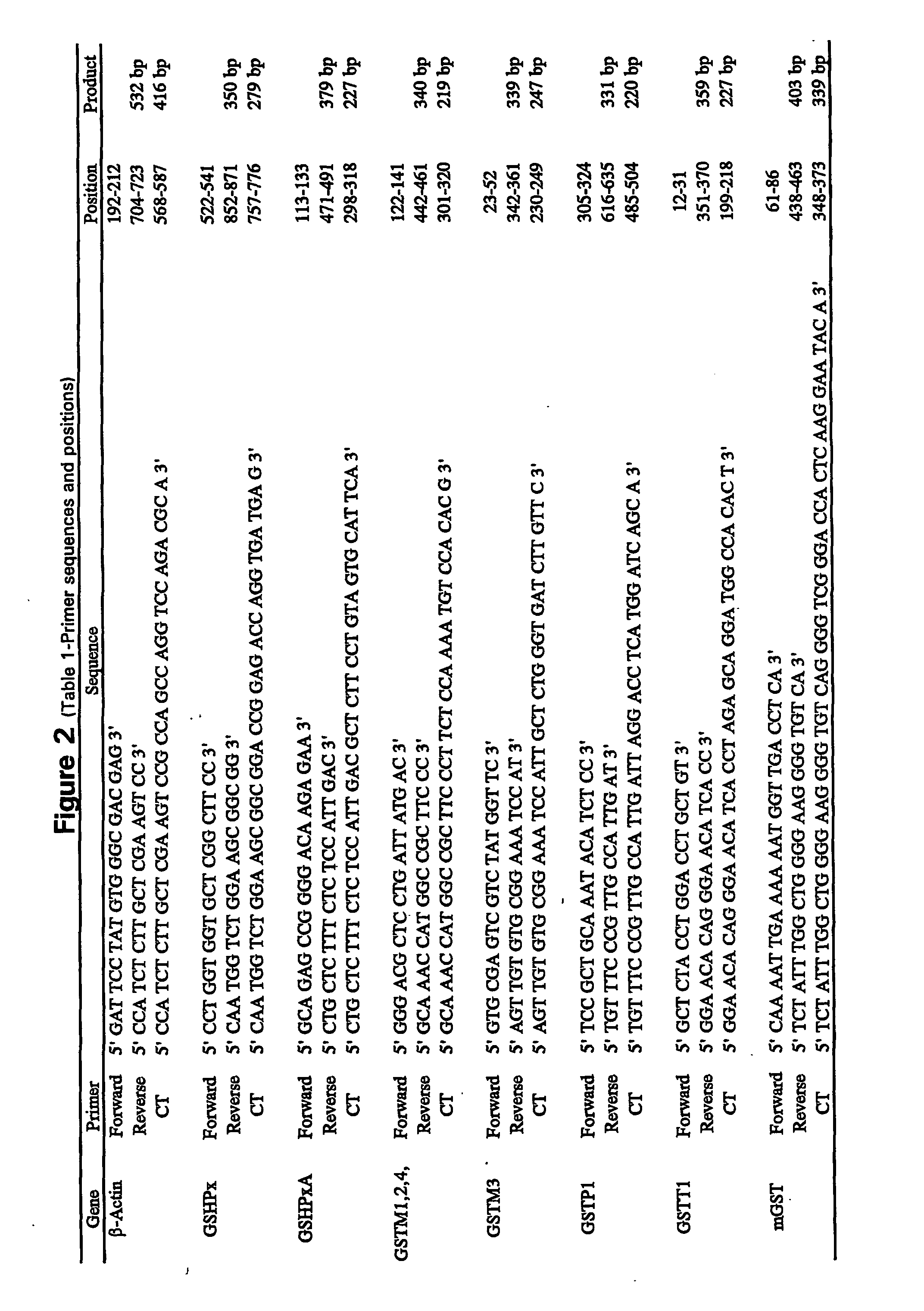
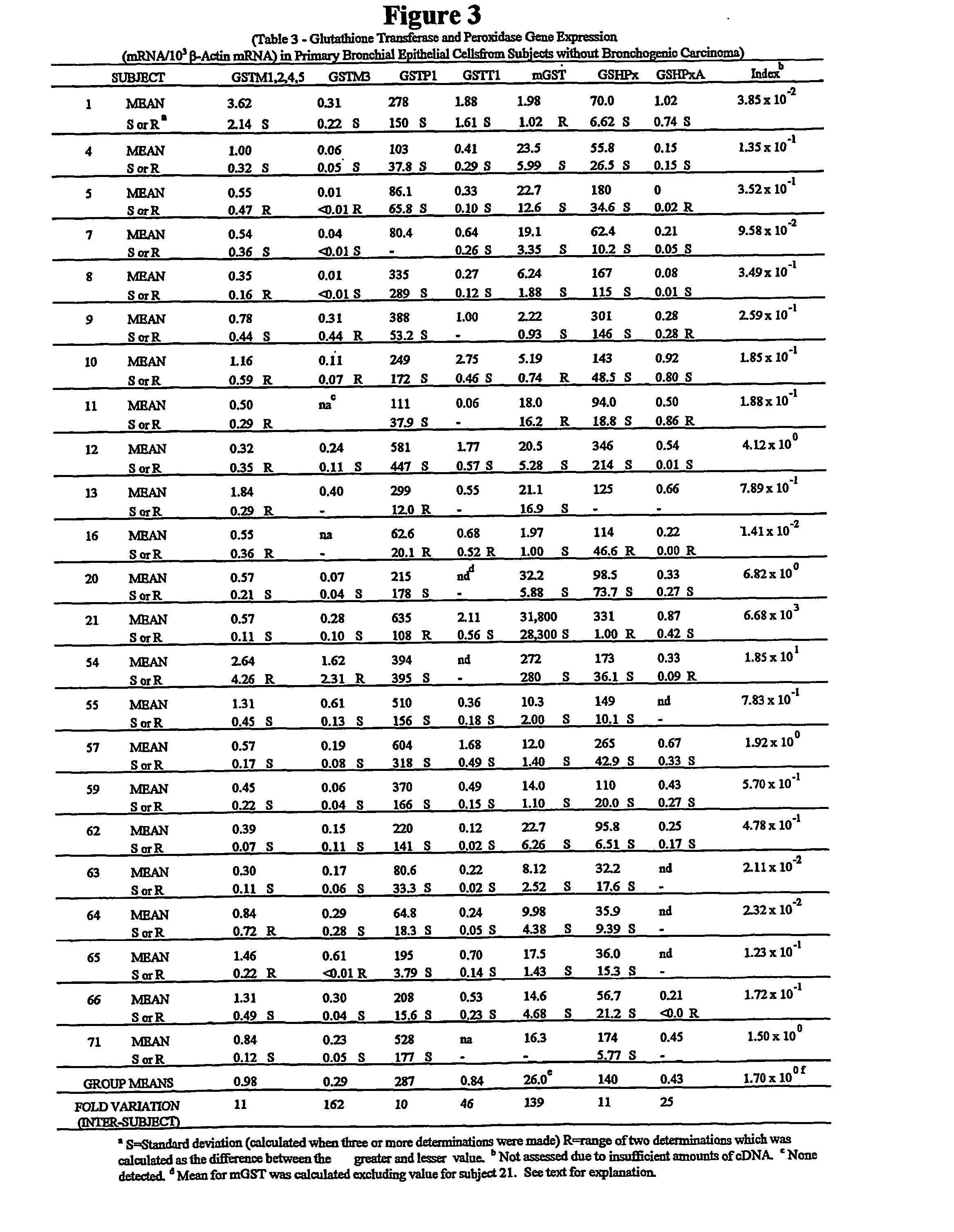
[0048] Reagents. 10.times.PCR buffer [500 mM Tris (pH 8.3), 2.5 mg / .mu.l BSA, 30 mM MgCl2] was obtained from Idaho Technology, Inc. (Idaho Falls, Id.). Taq polymerase (5 units / .mu.l), oligo dT primers, Rnasin (25 units / .mu.l), pGEM size marker, and dNTPs were obtained from Promega (Madison, Wis.). Moloney murine leukemia virus reverse transcriptase (200 units / .mu.l), 5.times. first strand buffer [250 mM Tris-HCl (pH 8.3), 375 mM KCl, 15 mM MgCl2, 50 mM DTT], and RNase-free water were obtained from Life Technologies, Inc. (Gaithersburg, Md.), NuSieve and SeaKem LE agarose were obtained from FMC BioProducts (Rockland, Me.). TriReagent was obtained from molecular Research Center (Cincinnati, Ohio), Bronchial epithelial cell growth medium was obtained from Clonetics (San Diego, Calif.). Natural human fibronectin and collagen (type 1 rat tail) were obtained from Collaborative Biomedical Products (Bedford, Mass.). All other chemicals and reagents were molecular biolog...
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| concentrations | aaaaa | aaaaa |
| density | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com