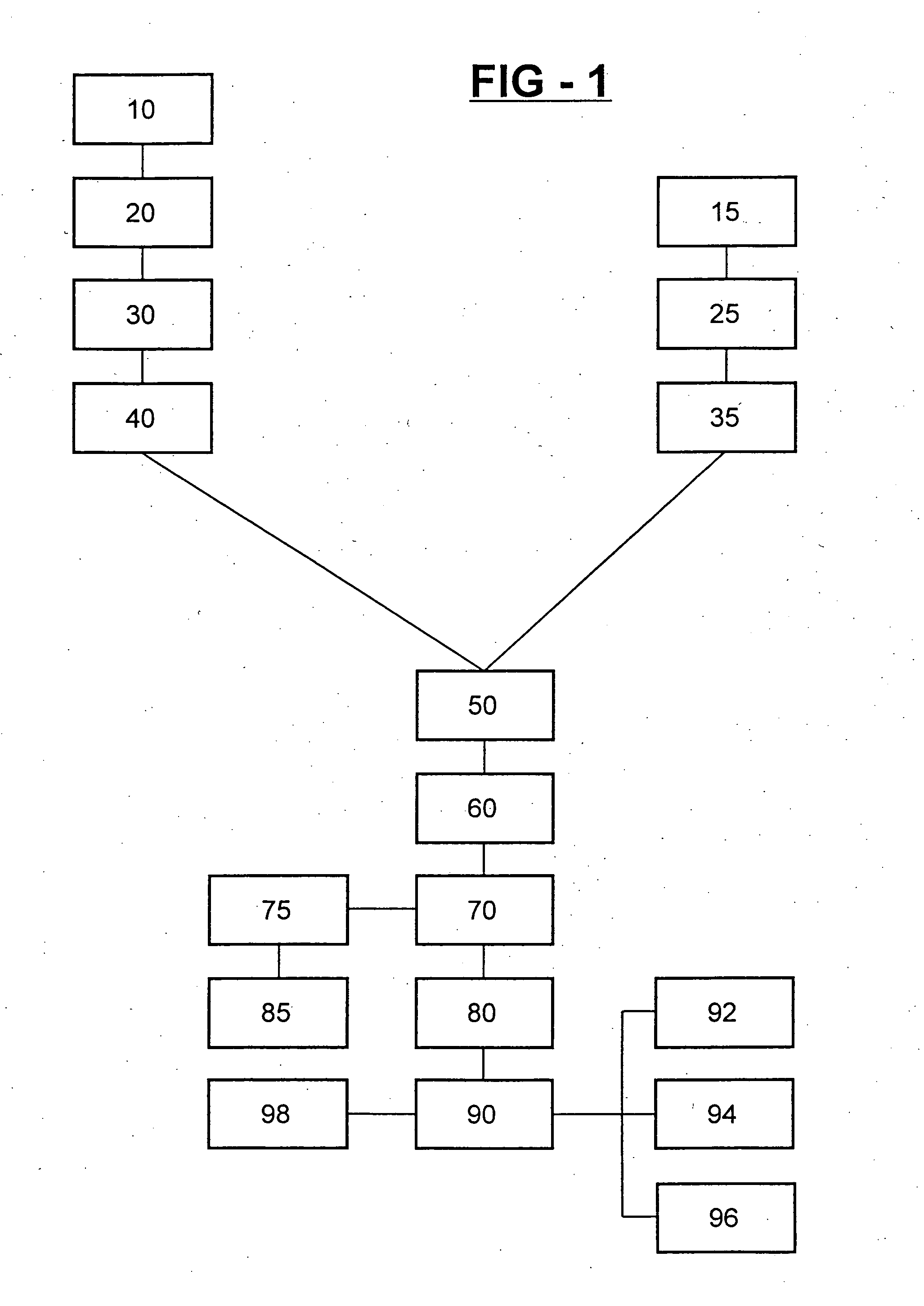
Cell-based method and assay for measuring the infectivity and drug sensitivity of immunodeficiency virus
a cell-based method and immunodeficiency virus technology, applied in the field of genetically modified cells, can solve the problems of impeded the development of useful approaches for detecting, quantifying and analysing hiv infection of primary virus isolates, and unable to achieve cost-effectiveness
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Generation of Transduction Vectors for the Delivery of Marker Genes
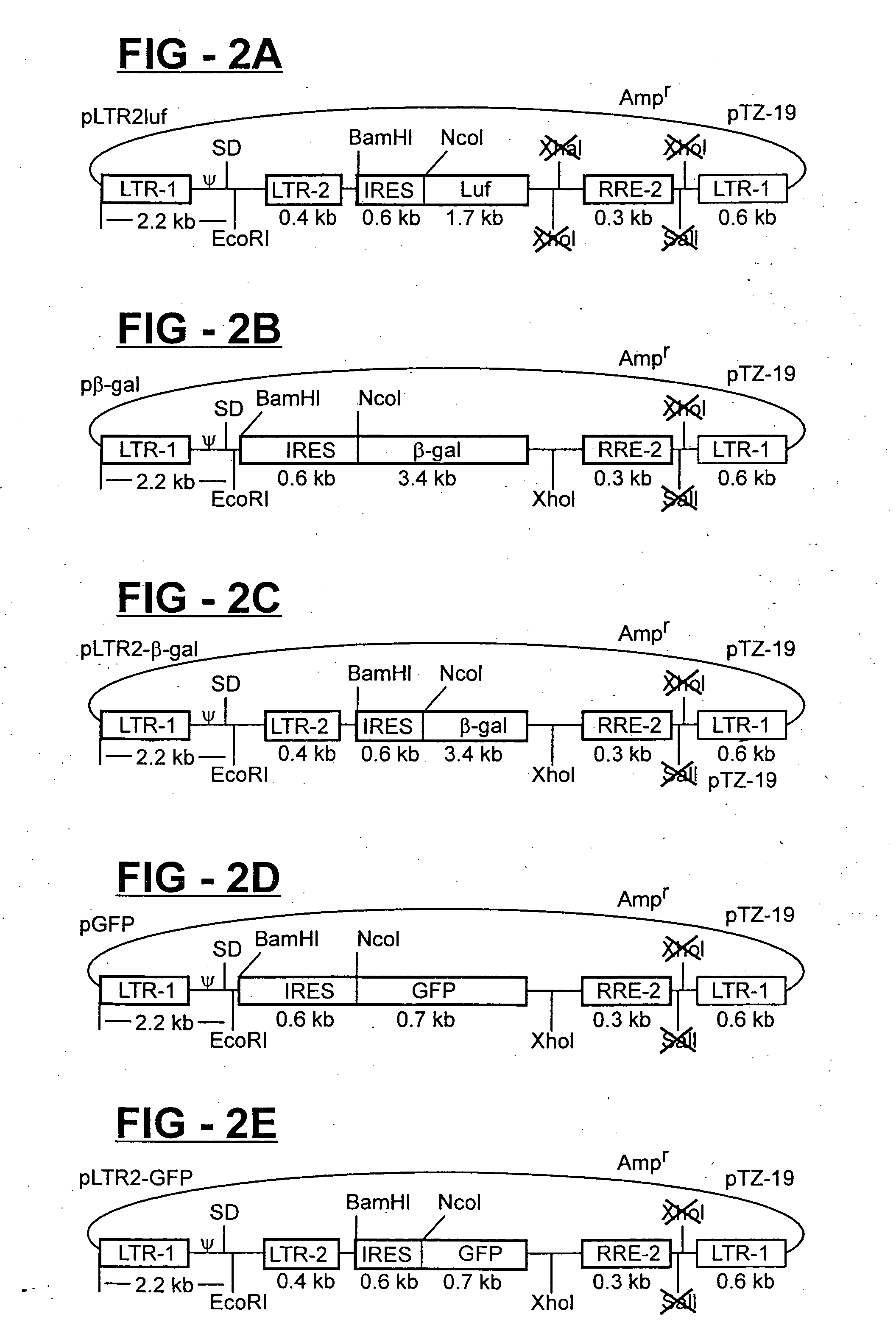
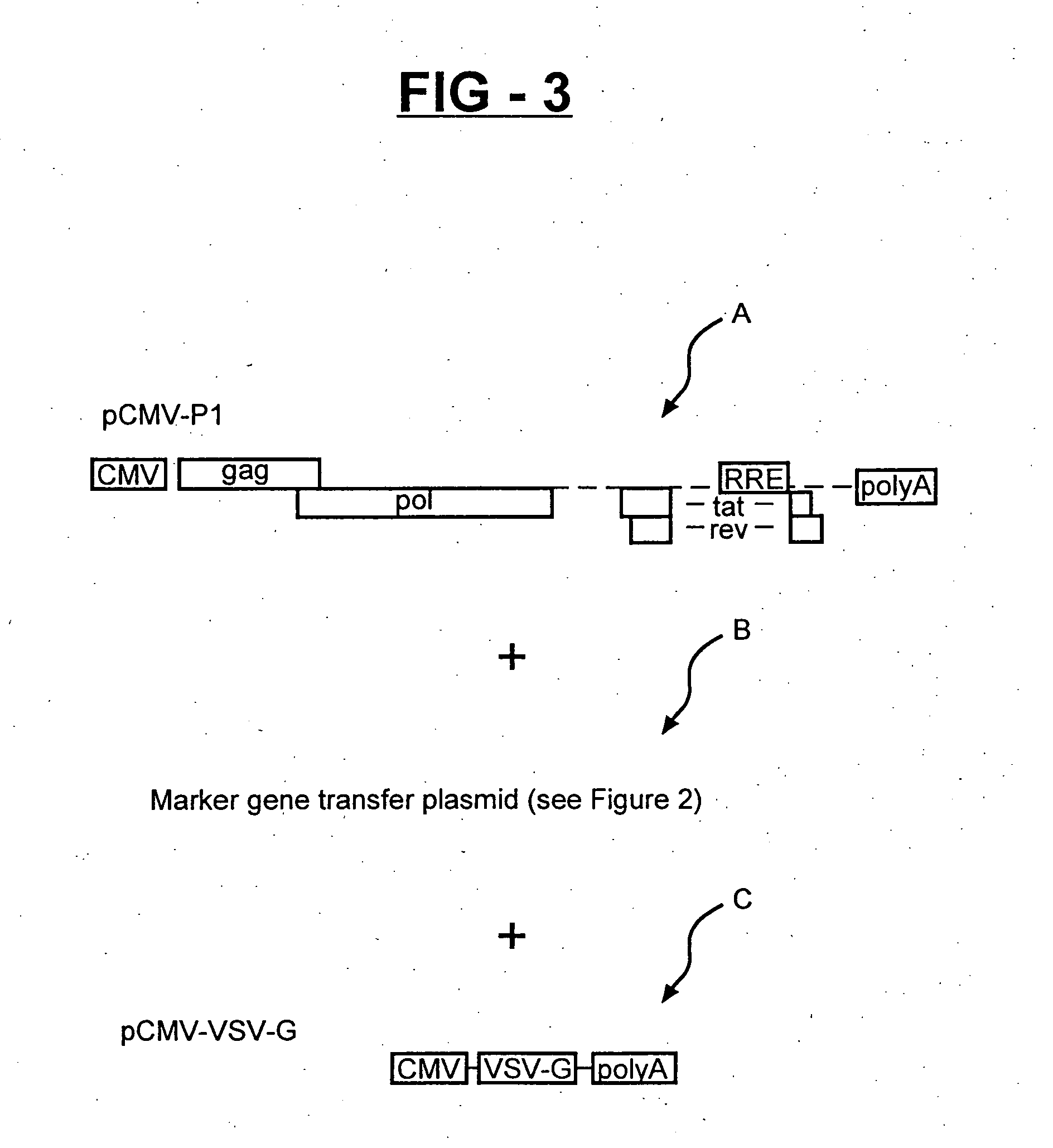
[0036] In order to generate vector stocks for transduction with the different reporter genes, the β-gal, GFP and luciferase gene transfer plasmids, including those containing, the HIV-2 LTR (shown in FIG. 2), are separately transfected into cultures of 293T cell together with a lentiviral-based packaging plasmid (pCMV-GP1), and the pCMV-VSV-G env plasmid (FIG. 3). Forty-eight hours later, the vector-containing culture supernatants are harvested, clarified by low-speed centrifugation, filtered through 0.45 micron filters, analyzed for HIV-1 p24 core antigen concentration by ELISA, aliquoted, and cryopreserved as stocks. Four serial five-fold dilutions (normalized for p24 antigen concentration) of the stocks are prepared and used to infect replica cultures of HIV-HeLa cell. The HIV-HeLa cells contained an integrated HIV-1 provirus that is defective in vpr and env, and produces the Tat and Rev protein for transactivati...
example 2
Generation of β-gal, Luciferase and GFP Indicator Cell Lines to Quantify HIV / SIV Infection
[0037] The following pairs of vector stocks (derived as described above) are used to co-transduce cultures of HeLa-CD4 cells: (a) pluf+pβ-gal, (b) pluf+pLTR2-βgal, (c) pluf+pGFP, (d) pluf+pLTR2-GFP, (e) pLTR-2-luf+pβ-gal, (f) pLTR2-luf+pLTR-2β-gal, (g) pLTR2-luf+pGFP, (h) pLTR2-luf+pLTR-2-GFP. Three days later, the cells are biologically cloned by limiting dilution in 48 well plates. Wells containing clonal cells (confirmed after initial plating by microscopy) are expanded into replica cultures. One replica culture set is infected with HIV-1 / SG3 and analyzed for marker gene expression (HFIV-1 infection provided Tat and Rev to activate marker gene expression) as described above. Expression positive cells cultures are identified, expanded and cryopreserved.
[0038] Since the expression of relatively few molecules of luciferase produces substantial luciferase activity levels, 36 non-HIV-1 infected...
example 3
Sensitive Detection of HIV-1 Primary Viruses Using β-gal and Luciferase Reporter Genes
[0039] The present invention utilizes a combination of a reporter assay system for sensitively and rapidly quantifying, infectious HIV-1 over a wide linear range with a cell line which is highly sensitive to infection with both M-tropic and T-cell tropic viruses. Transduction of the CD4-CCR5 positive J53 cell clone (Dr. David Kabat, Oregon Health Sciences University, Portland, Oreg.) with the pluf and pβ-gal expression vectors as described above. The pluf and pβ-gal transduced J53 cells (termed J53-βgal / luf) are infected with six different virus isolates (using four five-fold serial dilutions) that were unable to efficiently infect other CD4, CXCR4 expressing cell lines (P4 or Hi5) or a CD4, CXCR4 expressing cell line (MAGI) (see Table 2). Table 1 shows that all viruses, including the macrophage tropic YU2 clone, included as a control, are highly infectious in the J53β-gal / luf cell line.
[0040] To...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com