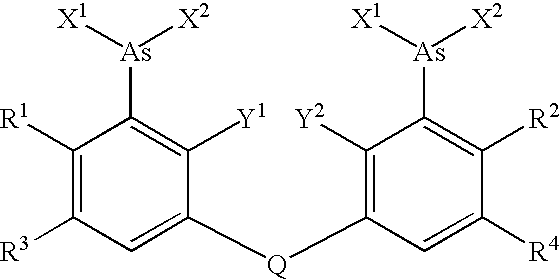
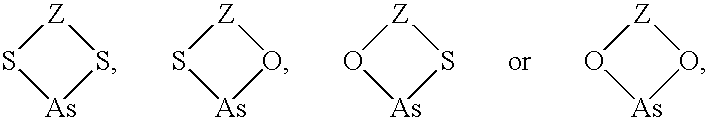
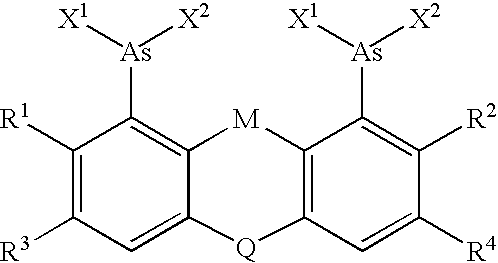
Target sequences for synthetic molecules
a synthetic molecule and target sequence technology, applied in the field of synthetic molecules, can solve the problems of labeling stoichiometry, inability to perform a large number of cells, and often encountered disruption of biological activity,
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Target Sequence Generated on AcpS
[0249] A target sequence that includes the SlyD (SEQ. ID NO: 4) tetracysteine sequence, CCGGKGNGGCGC (SEQ. ID NO: 5) was introduced onto the Carboxy-terminus of Acyl Carrier Protein S (AcpS). Since AcpS has only one endogenous cysteine amino acid and since AcpS is a robust stable protein, a substitution at the Carboxy-terminus could be made without altering the solubility of the properly folded protein. The four cysteines comprising the SlyD tetracysteine sequence were introduced at the carboxy-terminus of the protein as seen in SEQ. ID NO: 6. The mutated AcpS is referred to as AcpS+4Cys. The substitutions were generated using polymerase chain reaction with primers specific for the encoding the expression of the desired tetracysteine sequence. The nucleic acid sequence encoding the cysteine substituted AcpS was inserted into the pRSET vector (Invitrogen, Carlsbad, Calif., Catalog # V351-20) using restriction sites inherent to the vector's multiple c...
example 2
Binding Modes for Biarsenical Molecules to Target Sequences
[0251] The mode of binding of a biarsenical to a target sequence was examined using the Expressway™ in vitro protein synthesis kit (Invitrogen, Carlsbad, Calif.) and SDS-PAGE. Following the manufacture's protocol 1 μg of SlyD+His tag (SEQ. ID NO: 8), SlyD-C167A / C168A (SEQ. ID NO: 9), and SlyD-trunc171 (SEQ. ID NO: 10) vector DNAs were added to a total volume of 50 μL of S30 E. coli extract and reaction buffer. The reaction was placed at 37° C. with 225 rpm shaking for two hours. After incubation 5 μL of RNase A was added to the reaction, after which an additional 15 minute incubation at 37° C. was performed. Protein from the in vitro protein synthesis reaction was prepared for SDS-PAGE analysis through an acetone precipitation procedure. 5 μl of reaction was added to 20 μL of 100% acetone. After mixing well the acetone solution was centrifuged for 5 minutes at room temperature in a microcentrifuge at 12,000 rpm. The superna...
example 3
Specificity of Biarsenical Molecules for Tetracysteine Sequences
[0253] To demonstrate specificity of biarsenical compounds for different tetracysteine sequences several chimeric proteins were constructed. The native SlyD sequence (SEQ. ID NO: 4) was cloned into the pRSET vector (Invitrogen, Carlsbad, Calif.) using standard molecular biology techniques. Purified protein was produced from this vector by first transforming BL21 (DE3) cells (Invitrogen, Carlsbad, Calif., Catalog # C6010-03) and plated on LB-ampicillin plates. A single colony was selected and grown in one liter of liquid LB broth to a density of 1 O.D. and 1 mM IPTG was added to induce protein expression. After three hours of protein induction the culture was harvested by centrifugation at 10,000×g for 5 minutes at 4° C. The cell pellet was resuspended in 50 mM HEPES (pH 7.5), 140 mM NaCl and sonicated on ice for a total of two minutes. The E. coli lysate was separated by centrifugation at 25,000×g for 20 minutes at 4° ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| total volume | aaaaa | aaaaa |
| volume | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com