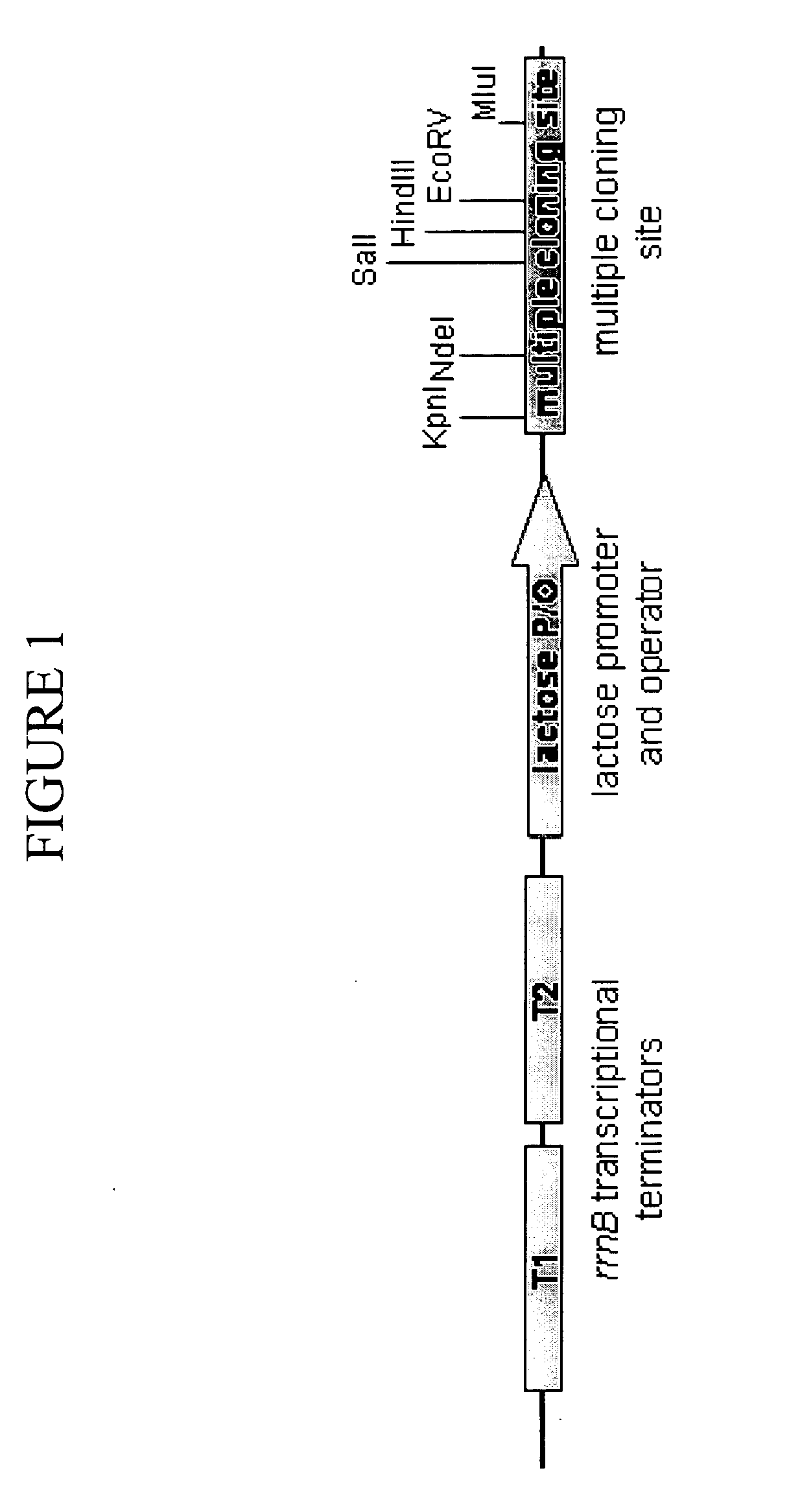
Systems for tightly regulated gene expression
a gene expression and tightly controlled technology, applied in the field of tightly controlled bacterial expression vectors, can solve the problems of leakage of expression systems, compromising the regulation of cloned inserts, and unwanted background expression of genes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Plasmid Construction
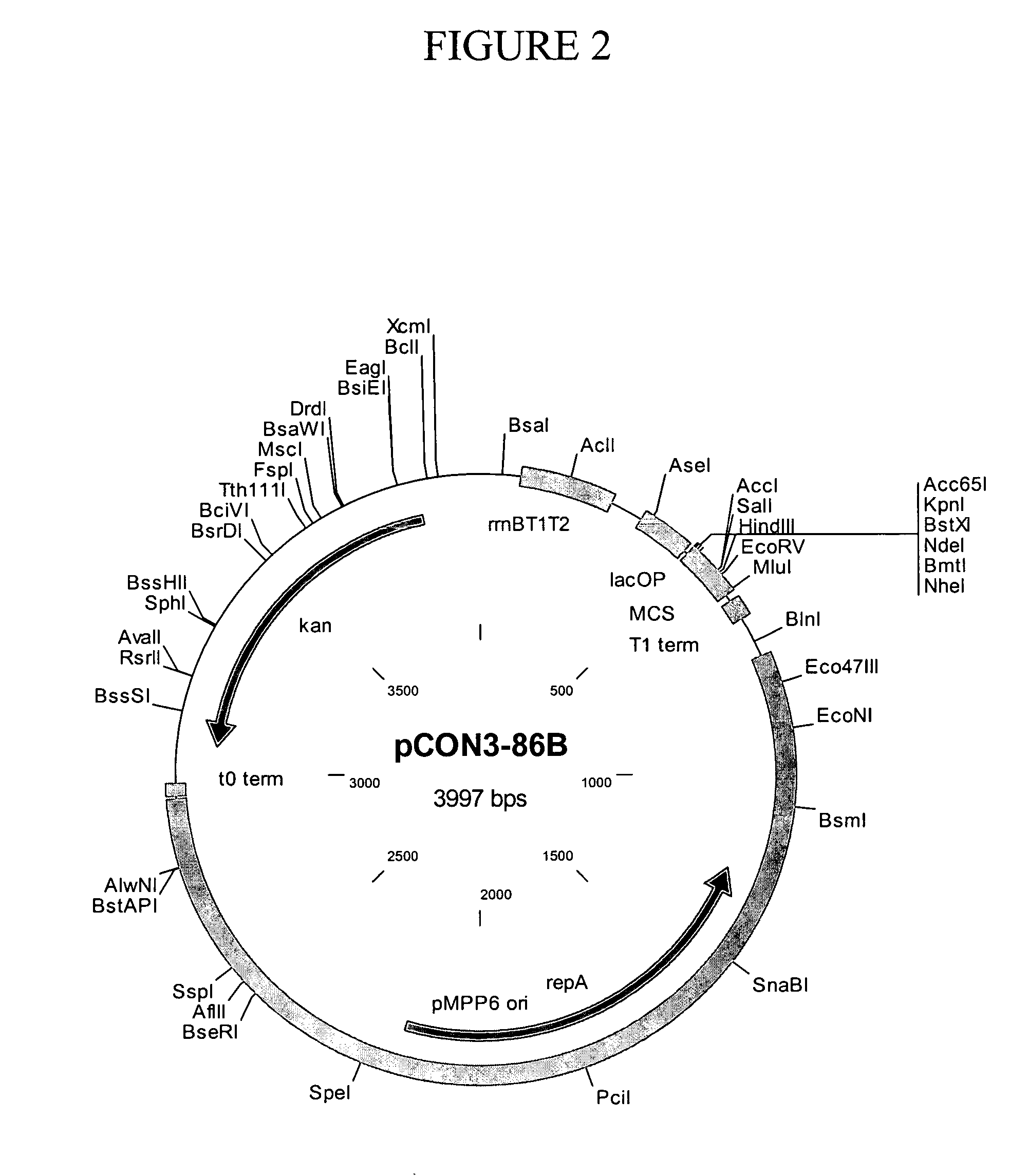
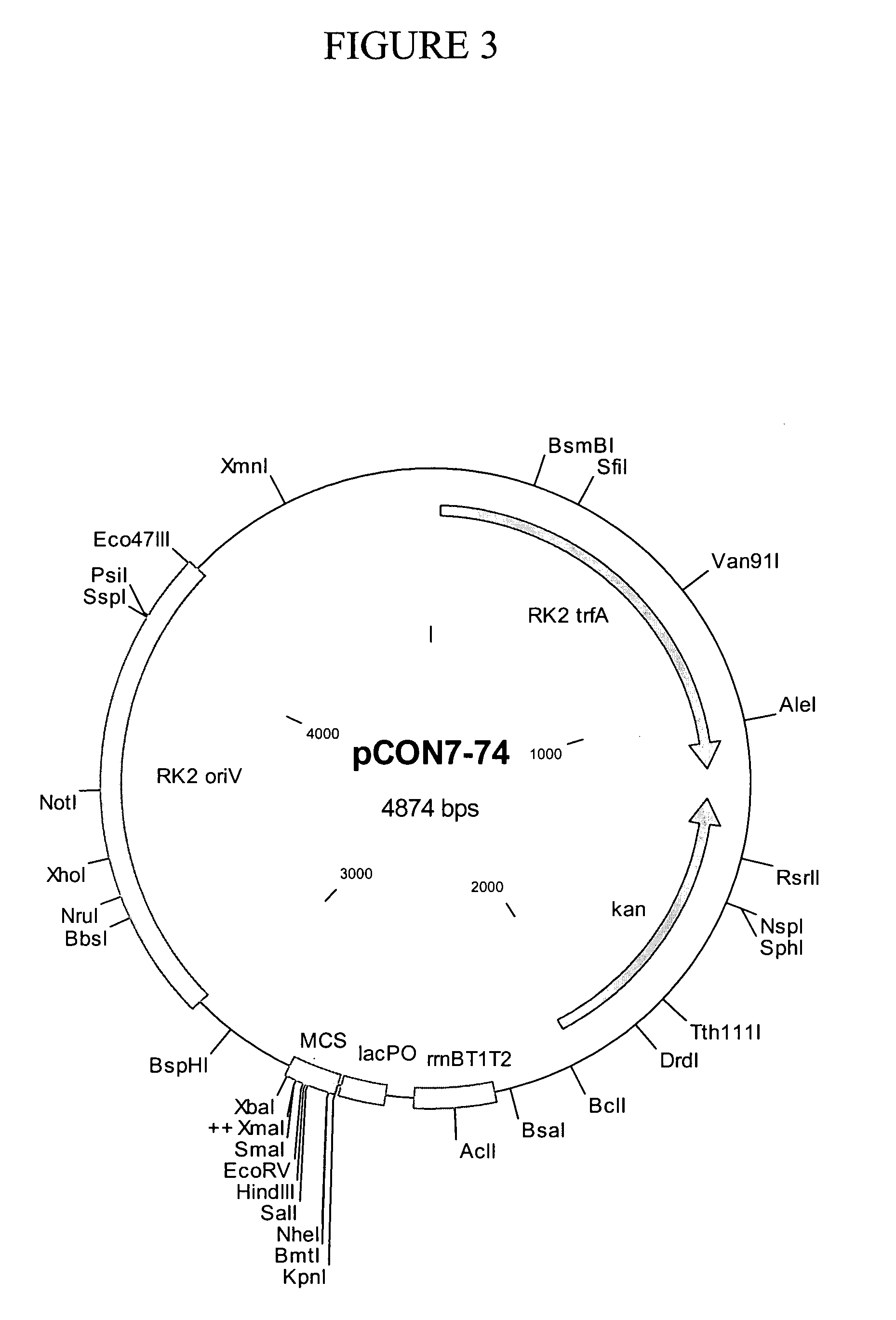
[0098] This Example describes the construction of exemplary plasmids of the present invention. Table 1 shows the names and corresponding Figure and SEQ ID NO designations for the plasmids described below. Sequences of plasmids and selected vector elements are shown in FIG. 11.
TABLE 1PlasmidsNameFigure (depicting map)SEQ ID NOpCON3-86B21pCON7-7432pCON7-7143pCON5-2554pCON7-7765pCON7-5876pCON4-4287pCON7-1198
A. Materials and Methods
Bacterial Strains and Media
[0099] The Escherichia coli strain utilized was NovaBlue {endA1 hsdR17(rK12-mK12+) supE44 thi-1 recA1 gyrA96 relA1 Δlac F′(proA+B+lacIqZAM15::Tn10 (TcR))} from Novagen (Madison, Wis.). All cloning was performed using standard methods known in the art, and using Luria Bertani growth media supplemented with 50 μg / ml kanamycin to permit selection for plasmids. For cloning of toxic gene products such as the colicins, the growth media was supplemented with 0.8% glucose to further repress the lactose promoter.
...
example 2
Gene Expression
[0108] This example describes the measurement of levels of expression from the vectors described in Example 1.
[0109] Using the standard assay for β-galactosidase activity, the promoter activity for vectors pCON3-86B, pCON5-25, pCON7-74, and pCON7-77 were obtained in repression conditions (Luria-Bertani broth supplemented with 0.8% glucose and 50 μg / ml kanamycin) and expression conditions (Luria-Bertani broth supplemented with 1 mM IPTG and 50 μg / ml kanamycin). Cultures were assayed in duplicate at an OD600 nm of 0.3-0.5 and expressed as Miller Units. The results are shown in FIG. 10.
[0110] As observed in FIG. 10, the promoter activities of pCON5-25 and pCON7-77 in repression medium are not significantly different from vectors pCON3-86B and pCON7-74, which do not contain the gene for α-galactosidase. However, upon de-repression with 1 mM IPTG, the promoter activity of pCON5-25 (with modified-pSC101 origin) is increased approximately 50-fold and the activity of pCON7...
example 3
Expression of Toxic Proteins
[0111] The vectors of the present invention were used to clone and stably maintain the genes encoding colicins D (pCON7-58), E3 (pCON4-42), E7 (pCON7-11), E3 (pCON12-82) in the absence of the cognate immunity proteins, with the ability to achieve high levels of protein / RNA expression upon de-repression of the promoter.
PUM
| Property | Measurement | Unit |
|---|---|---|
| Nucleic acid sequence | aaaaa | aaaaa |
| Toxicity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com