Double stranded linear nucleic acid probe and uses thereof
a nucleic acid probe and linear technology, applied in the field of nucleic acid amplification and detection, can solve the problems of unsatisfactory reaction kinetics of probes, prone to rapid mutation of viral rna targets in the bodies of hosts, and unsatisfactory for the detection of viral nucleic acids
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
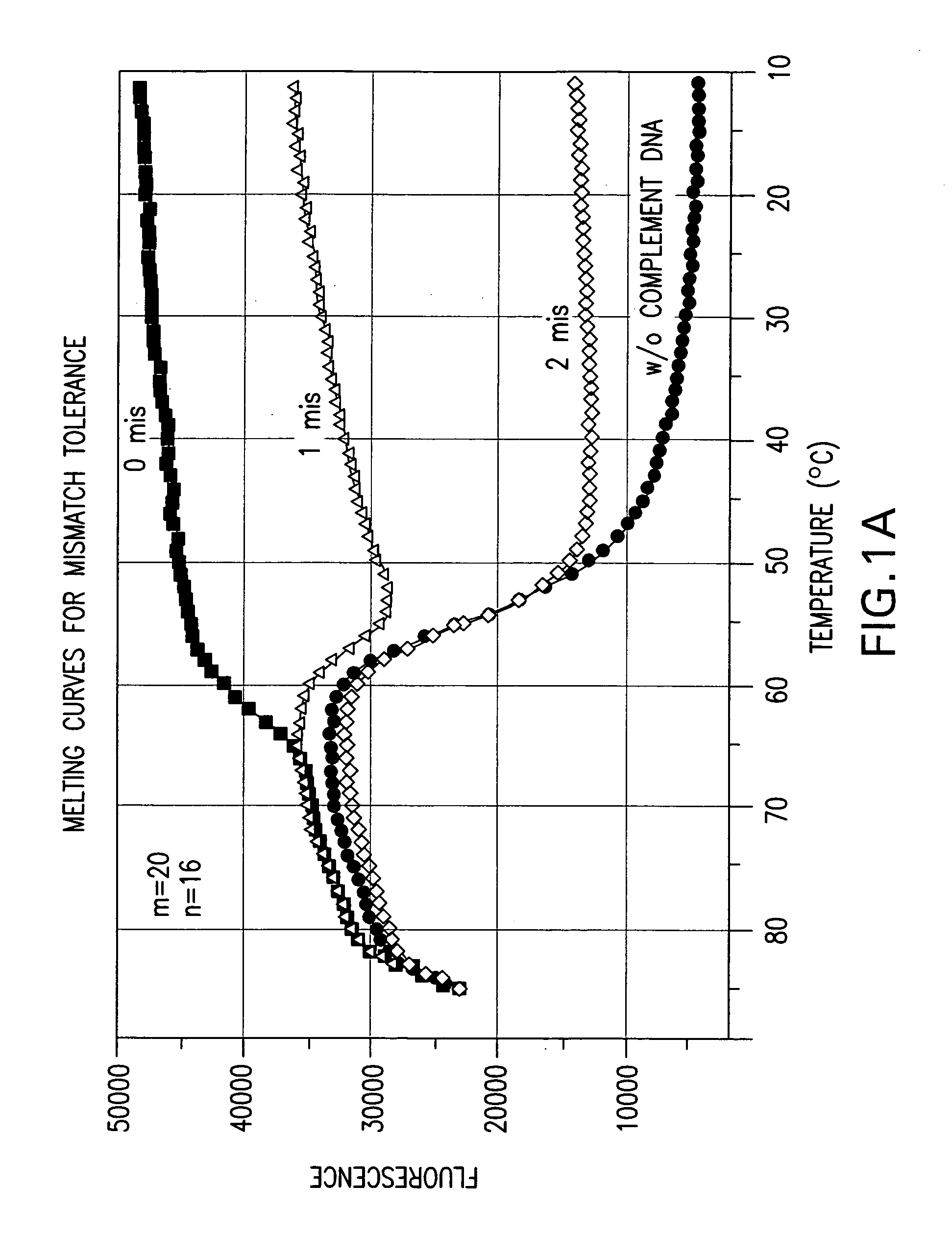
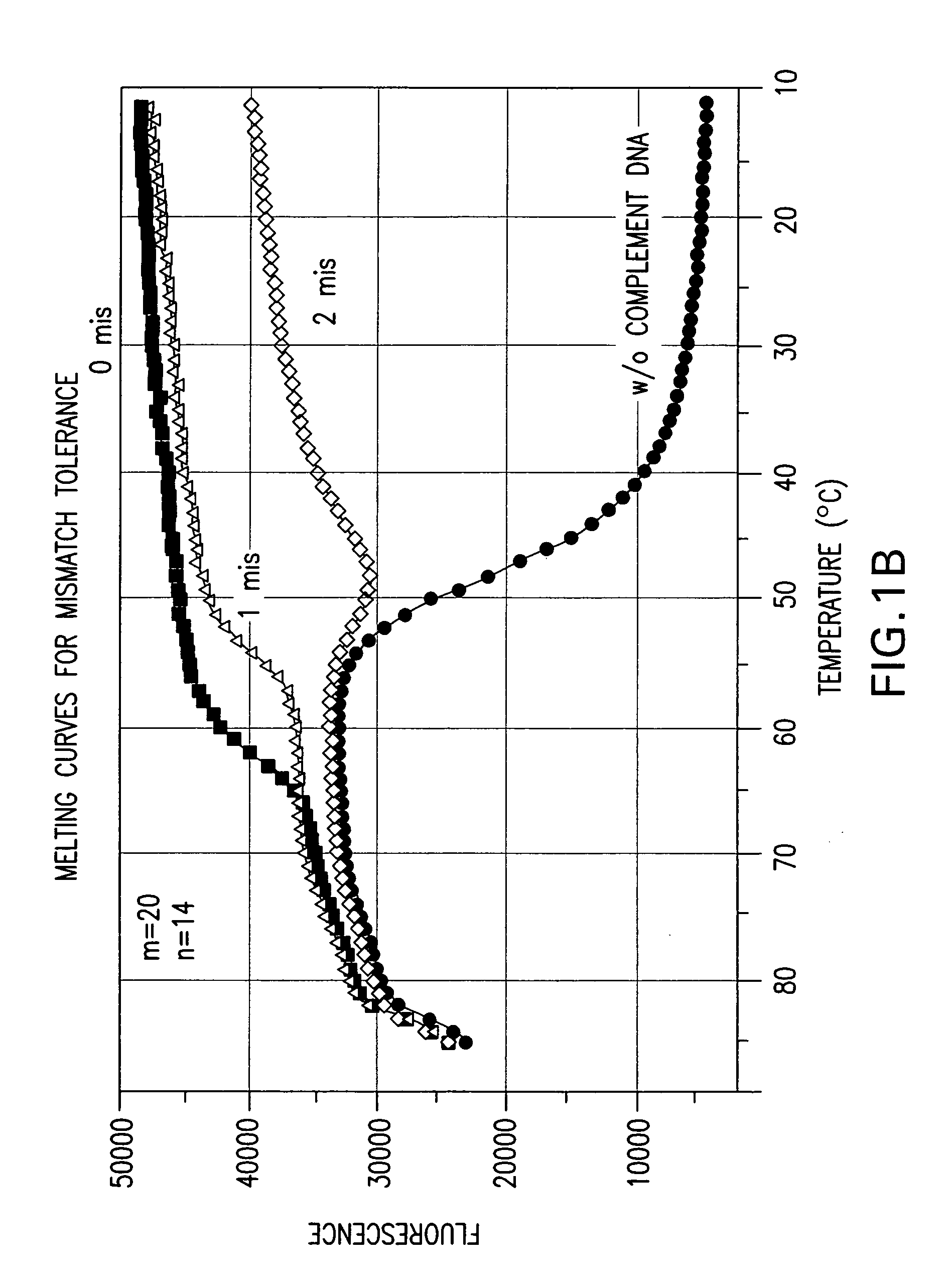
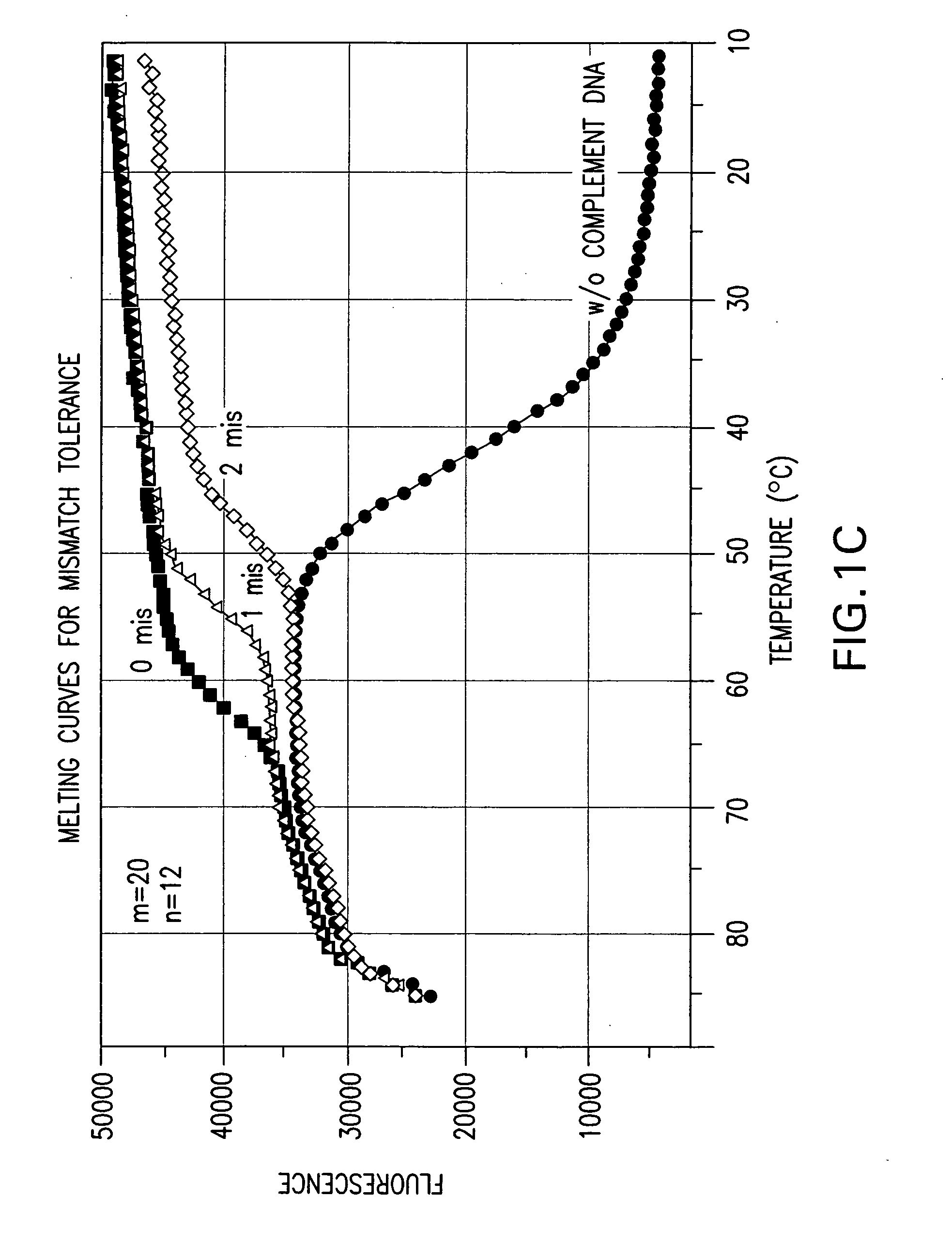
Effect of the Length Difference Between the Two Oligonucleic Acids of a Nucleic Acid Probe on Mismatch Tolerance Evaluated by Melting Curve Assays
[0056] Melting reactions were performed in a Stratagene Mx4000 multiplex quantitative PCR system with the following cycle conditions: 1 cycle of denaturation at 95° C. for 3 min; 75 cycles of 1-minute holding at a range of temperatures from 85° C. to 10° C. with an 1° C. decrement per cycle. Fluorescein (FAM) fluorescence measurements were recorded during each 1-minute hold of the 75 cycles. At the end of each run, the data were analyzed and melting curves were generated.
[0057] Table 2 sets forth the sequences of PCR primers and linear probes used in this and the following examples.
TABLE 2NameSequencePCR PrimersFP-295′ - ATTCCCTACAATCCCCAAAGTCAAGGAGT - 3′(SEQ ID NO:1)RP-255′ - CCCCTGCACTGTACCCCCCAATCCC - 3′(SEQ ID NO:2)RP-245′ - CCCCTGCACTGTACCCCCCAATCC - 3′(SEQ ID NO:3)Linear Probes1 labeled with FAM520-206-FAM- (5′) - ACAGCAGTACAAATG...
example 2
Effect of the Length Difference Between the Two Oligonucleic Acids of a Nucleic Acid Probe on Mismatch Tolerance Evaluated by Quantitative Real-Time RT-PCR Assays
[0060] This example shows the evaluations on three different probe sets 520-20 / que-16, 520-20 / que-12 and 520-31 / que-14 for their mismatch tolerances by performing the quantitative real-time RT (reverse transcription)-PCR assays. Five transcripts carrying different mutations were employed to test these three probe sets. In these non-competitive quantitative assays, each 100 μl RT-PCR reaction contained 1.25×RT-PCR buffer (62.5 mM Bicine, pH 8.05-8.25, 143.75 mM potassium acetate, 10% glycerol, 0.125 mM EDTA, 0.0125 mg / ml acetyl bovine serum albumin (BSA), 0.078% (v / v) Tween 20, and 0.025% (w / v) sodium azide), 2.5 mM MnCl2, 0.375 mM of each deoxynucleotide-triphosphate (dATP, dCTP, dGTP, dTTP), 13.13 units of rTth DNA polymerase (Applied Biosystems), 0.6 μM HIV forward PCR primer FP-29, 1.6 μM HIV reverse PCR primer RP-25 (T...
example 3
Inhibition of RT-PCR by a Quenching Oligonucleotide with a Tm Higher than the RT Temperature
[0063] To examine the impact of utilizing a quencher probe with a Tm that is above the incubation temperature of the RT reaction, performance of the probe combination, lin-41 and que-23 (Table 2), was evaluated in a quantitative real-time reverse transcription (RT)-PCR assay. The quenching oligo, que-23, has a Tm of 60.27, above the 59° C. RT incubation temperature. In this competitive quantitative assay, each 100 μl RT-PCR reaction was carried out in the presence of 1×EZ buffer (containing 50 mM Bicine, pH 8.2, 115 mM potassium acetate and 8% glycerol), 2.5 mM Mn(OAc)2, 0.4 mM of each deoxynucleotide-triphosphate (dATP, dCTP, dGTP, dTTP), 20 units of RNase inhibitor, 10 units of Tth DNA polymerase (all from Applied Biosystems), 0.2 μM HIV forward PCR primer FP-29, 1.0 μM HIV reverse PCR primer RP-24, 0.1 μM FAM-labeled oligo probe lin-41 for detection of the HIV wild type PCR products, 0.2 ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Ratio | aaaaa | aaaaa |
| Fluorescence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com