Methods for reducing the range in concentrations of analyte species in a sample
a technology of analyte and concentration range, which is applied in the field of combinatorial chemistry, protein chemistry and biochemistry, can solve the problems of inability to detect analyte species, inability to accurately measure the amount, and interference with the ability to detect less abundant analytes, so as to maintain the diversity of the population of analyte species
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
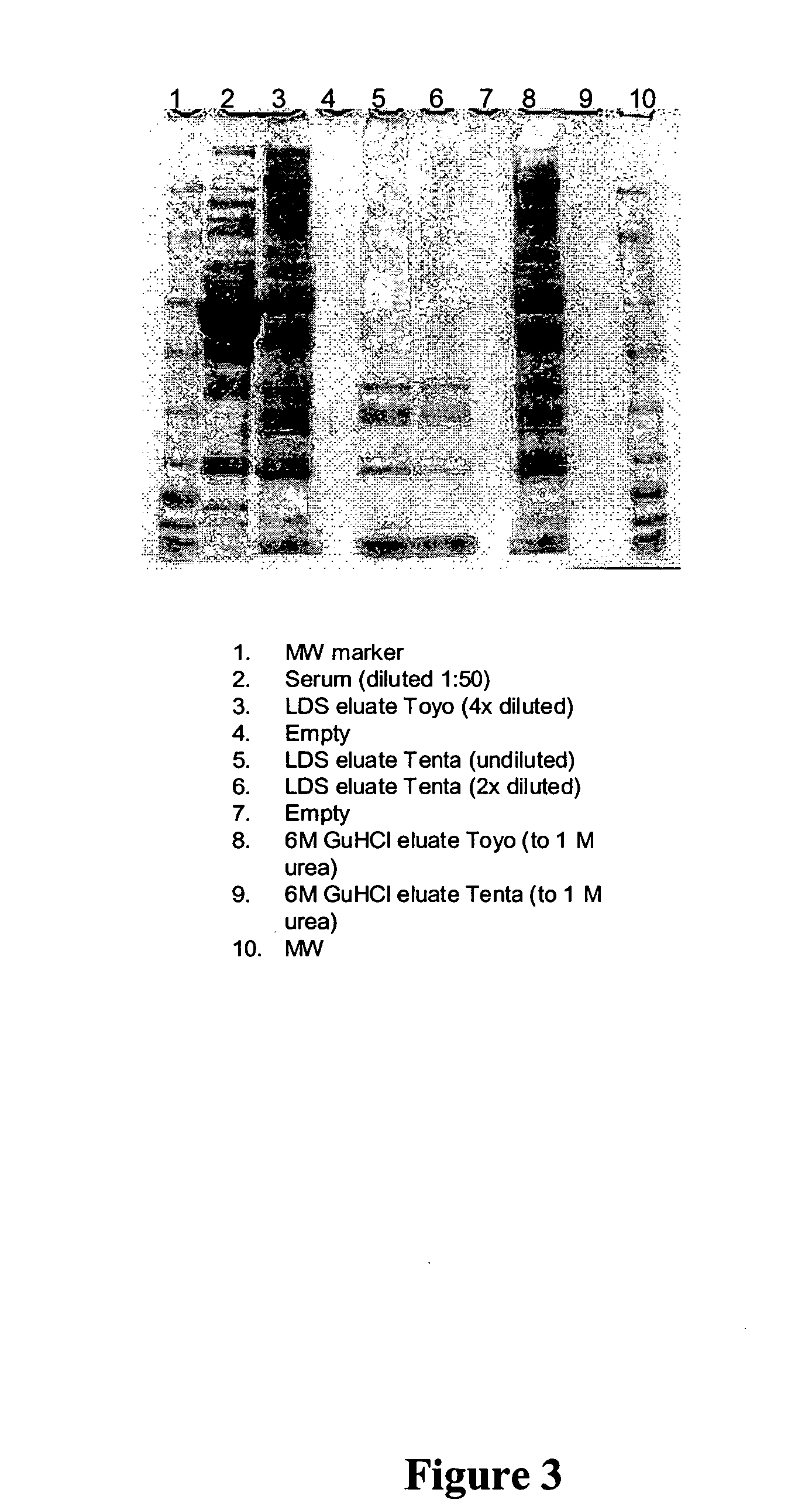
Reduction of Range of Concentrations of Human Serum Proteins
[0172] This example illustrates how one embodiment of the invention described above may be applied to a complex biological sample, in this case human serum. In this example, a reduction in the variance of serum protein concentrations is achieved by selectively adsorbing serum proteins to hexapeptides coupled to insoluble beads. More than 1×106 possible permutations of hexapeptide are represented in the binding moiety population of the example, in the form of a combinatorial library of split, recombine and pool beads. In this format, high abundance serum analytes, such as albumin, are bound to a hexapeptide binding moiety, but only to a level equal to saturation of the particular binding moiety. In contrast, low abundance serum analytes are bound almost in their entirety, as the amount of binding moiety recognizing the low abundance analyte is not limiting. The result of this selective binding is a reduction in the range of...
example 2
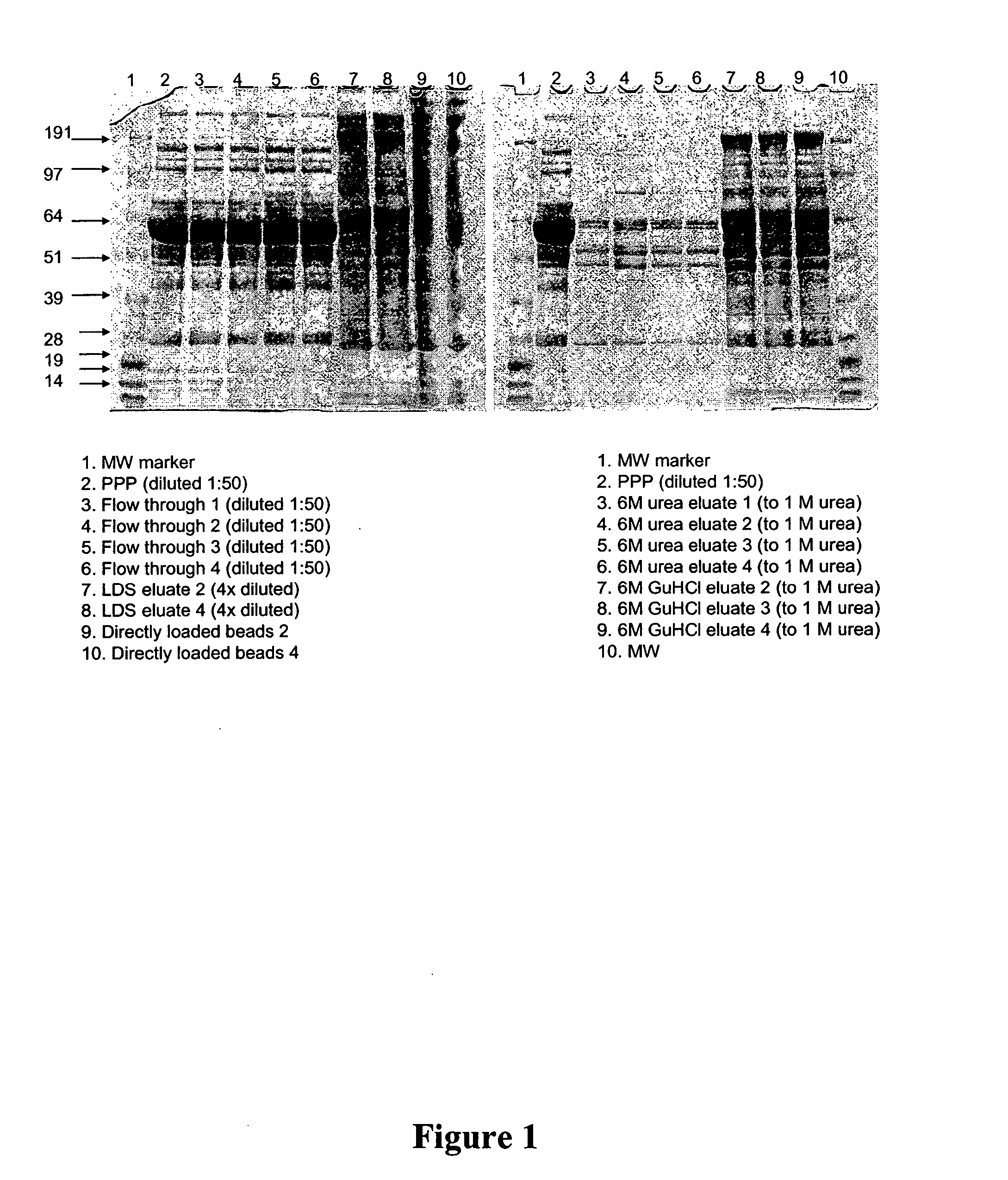
Incubation of Library with Unfractionated, Undiluted Human Pooled Plasma
[0177] To aid analysis of complex samples, this method is useful to decrease the concentration differential. Human plasma is one of the most complex and difficult to analyze materials: proteins are present in concentration range greater than 1010 (Anderson and Anderson); decreasing this range will aid in the analysis of trace proteins. Under the conditions of this method, incubation of plasma with the ligand library will increase the number of proteins that can be detected and subsequently analyzed as compared with analysis of the unprocessed starting material.
[0178] A. Sample Preparation
[0179] Frozen, pooled, human platelet-poor plasma (PPP) was thawed at 37° C. and filtered through 0.8 and 0.45 μm filters. Four replicates of approximately 1 ml of a library of hexamer peptide ligands on Toyopearl 650 M amino resin (65 μm average diameter, ˜2×106 beads / ml; Tosoh Biosciences, Montgomeryville, Pa.) with EACA-Al...
example 3
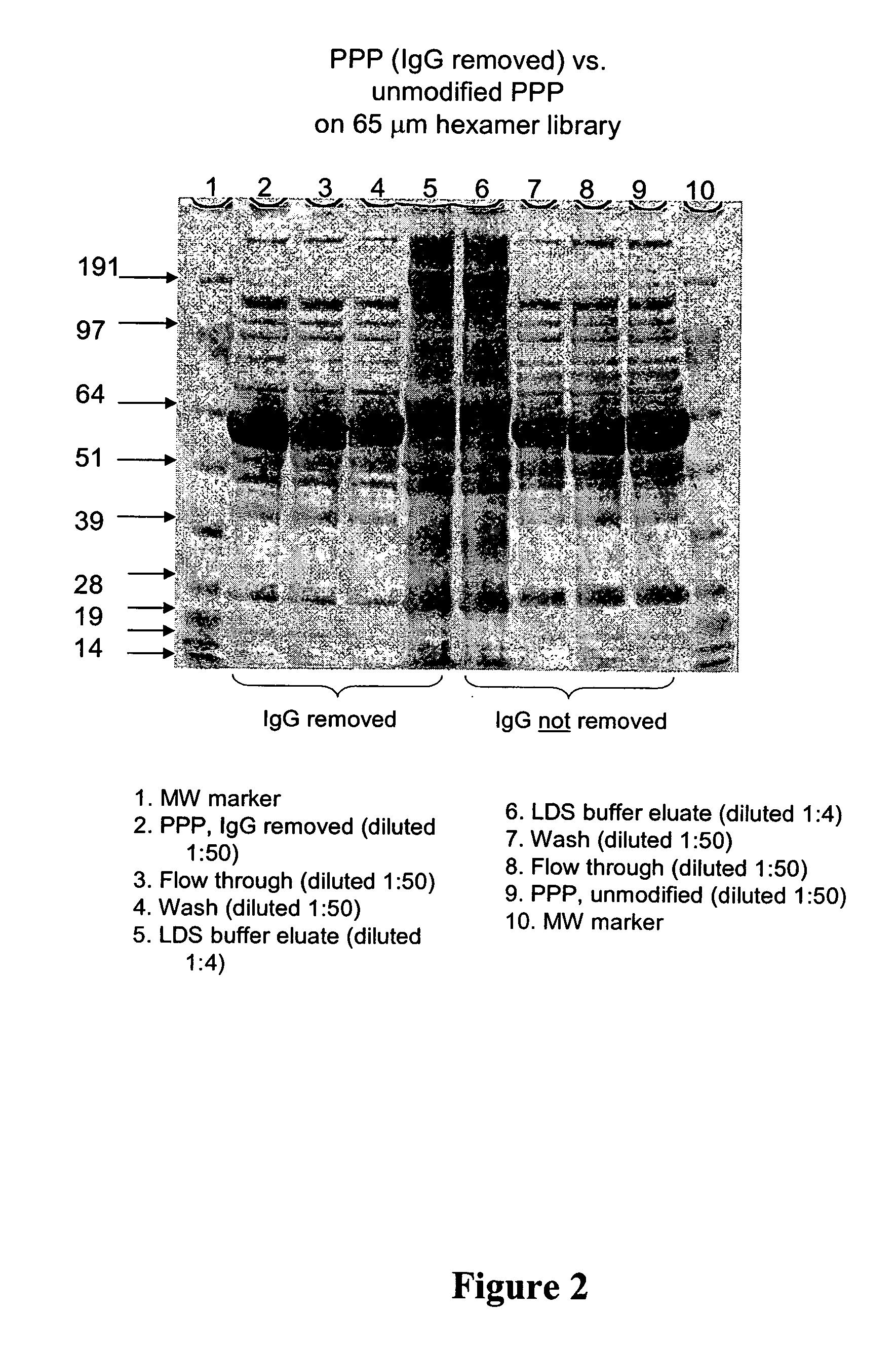
Reduction of Concentration Variance After Removal of IgG.
[0185] In many proteomic applications, one of the first steps of sample preparation is removal of albumin and IgGs, as these high abundance proteins mask the detection of lower abundance species. Removal of these proteins, however, also often removes trace species associated with them, and also involves loss of sample. It would be advantageous to have a method of sample preparation that does not require IgG depletion before analysis. This example demonstrates that removal of IgGs is not required to visualize protein species that are not detected in intact plasma. The pattern of proteins detected in LDS-PAGE is compared in plasma that has and has not been depleted of IgGs.
[0186] A. Sample Preparation
[0187] Frozen, pooled, human platelet-poor plasma (PPP) was thawed at 37° C. and filtered through 0.8 and 0.45 μm filters. IgG was removed from the plasma as follows: 5 ml Protein G Sepharose Fast-flow resin (Amersham, T&S) was p...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Diameter | aaaaa | aaaaa |
| Mass | aaaaa | aaaaa |
| Power | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com