Enrichment method for variant proteins with altered binding properties
a technology of binding properties and enrichment methods, which is applied in the field of systematic selection of novel binding proteins, can solve the problems of difficult methods for making non-naturally occurring synthetic binding partners, high cost and difficulty, and achieve the effect of high affinity binding proteins and efficient selection
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
examples
[0105] Without further description, it is believed that one of ordinary skill in the art can, using the preceding description and illustrative examples, make and utilize the present invention to the fullest extent. The following working examples therefore specifically point out preferred embodiments of the present invention, and are not to be construed as limiting in any way of the remainder of the disclosure.
example i
Plasmid Constructions and Preparation of hGH-Phagemid Particles
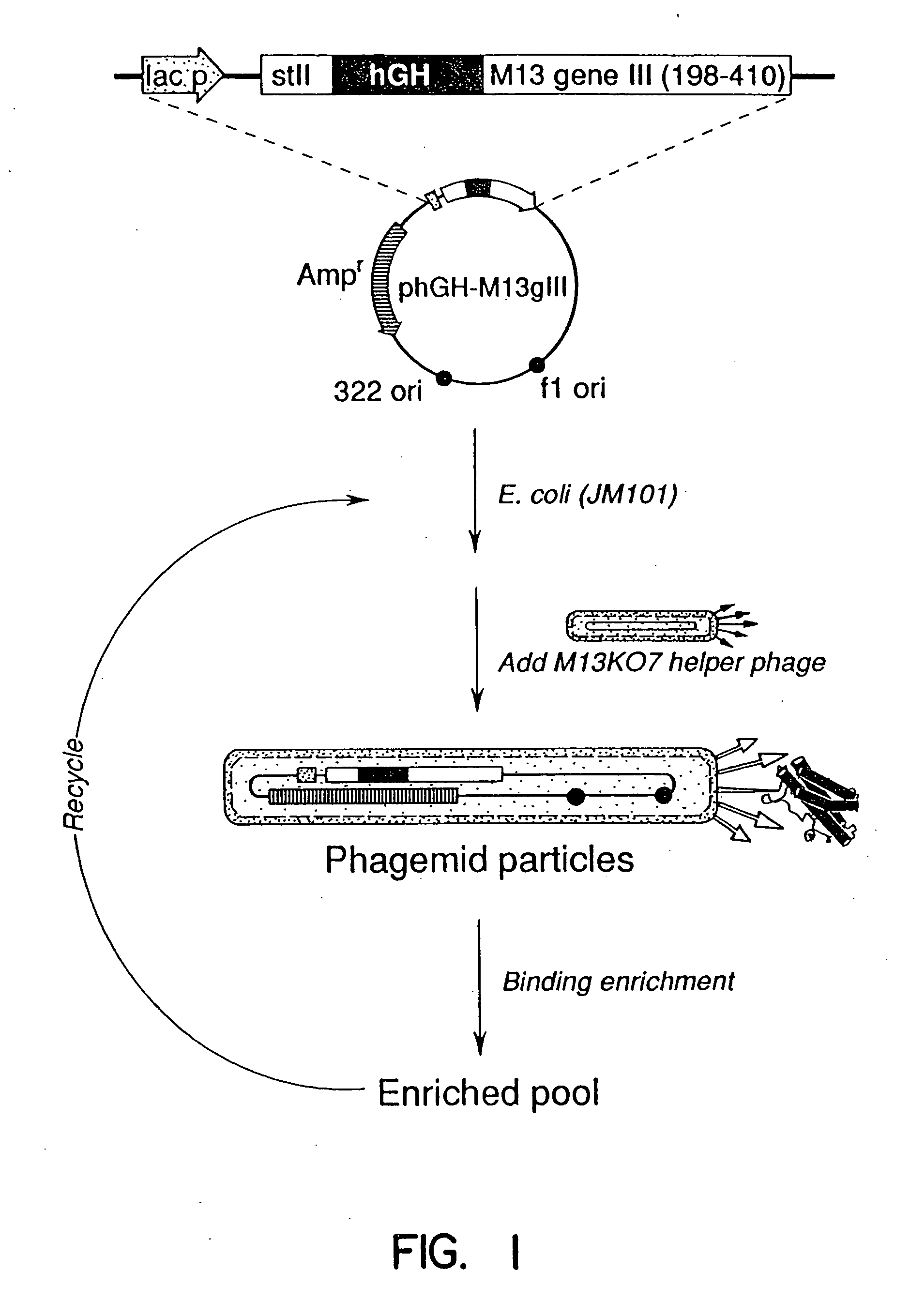
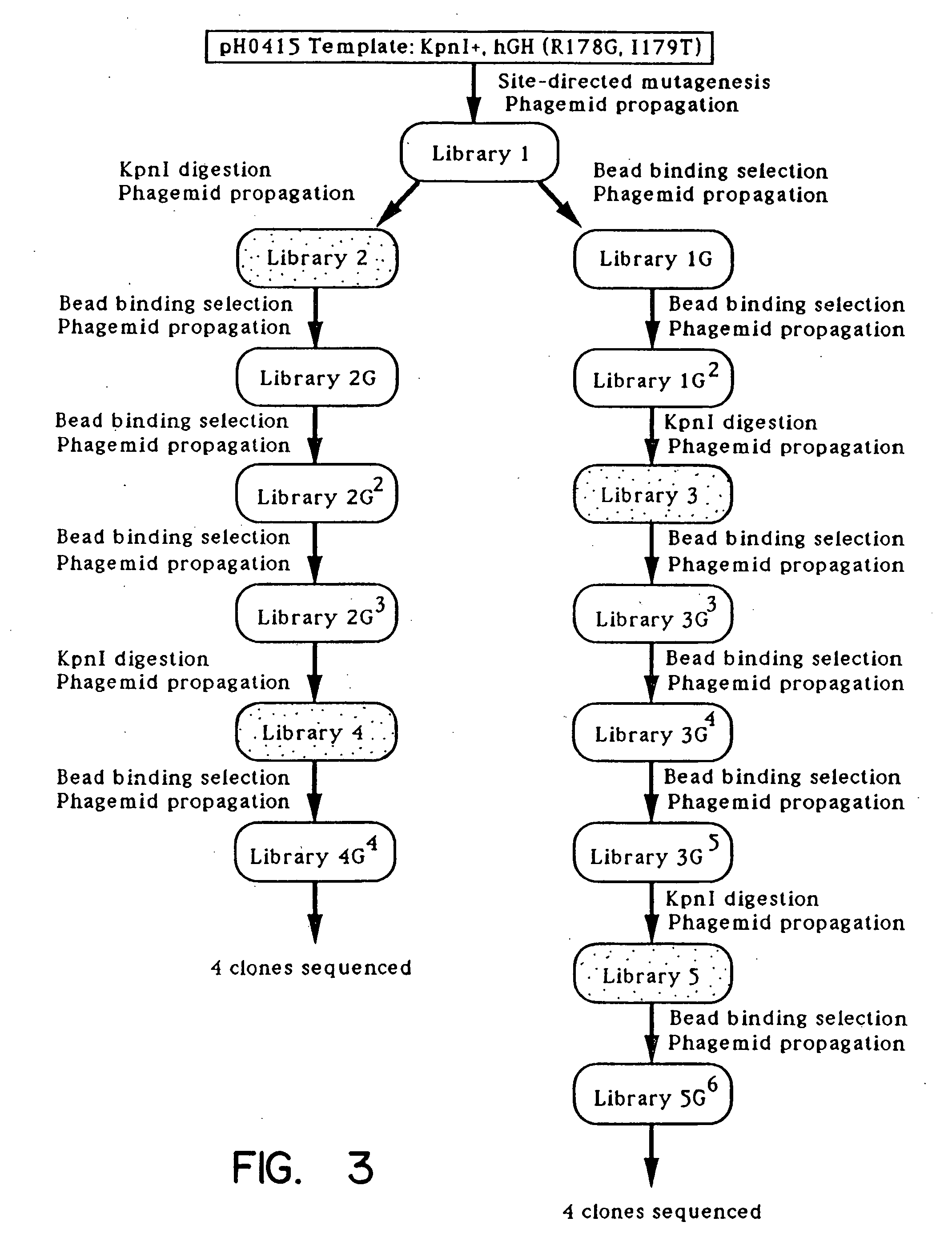
[0106] The plasmid phGH-M13gIII (FIG. 1), was constructed from M13KO77 and the hGH producing plasmid, pBO473 (Cunningham, B. C., et al., Science, 243:1330-1336, [1989]). A synthetic oligonucleotide 5′-AGC-TGT-GGC-TTC-GGG-CCC-TTA-GCA-TTT-AAT-GCG-GTA-3′ (SEQ ID NO:2) was used to introduce a unique ApaI restriction site (underlined) into pBO473 after the final Phe191 codon of hGH. The oligonucleotide 5′-TTC-ACA-AAC-GAA-GGG-CCC-CTA-ATT-AAA-GCC-AGA-3′ (SEQ ID NO:3) was used to introduce a unique ApaI restriction site (underlined), and a Glu197-to-amber stop codon (bold lettering) into M13KO7 gene III. The oligonucleotide 5′-CAA-TAA-TAA-CGG-GCT-AGC-CAA-AAG-AAC-TGG-3′ (SEQ ID NO:4) introduces a unique NheI site (underlined) after the 3′ end of the gene III coding sequence. The resulting 650 base pair (bp) ApaI-NheI fragment from the doubly mutated M13KO7 gene III was cloned into the large ApaI-NheI fragment of pBO473 to create...
example ii
Immunochemical Analyses of hGH on the Fusion Phage
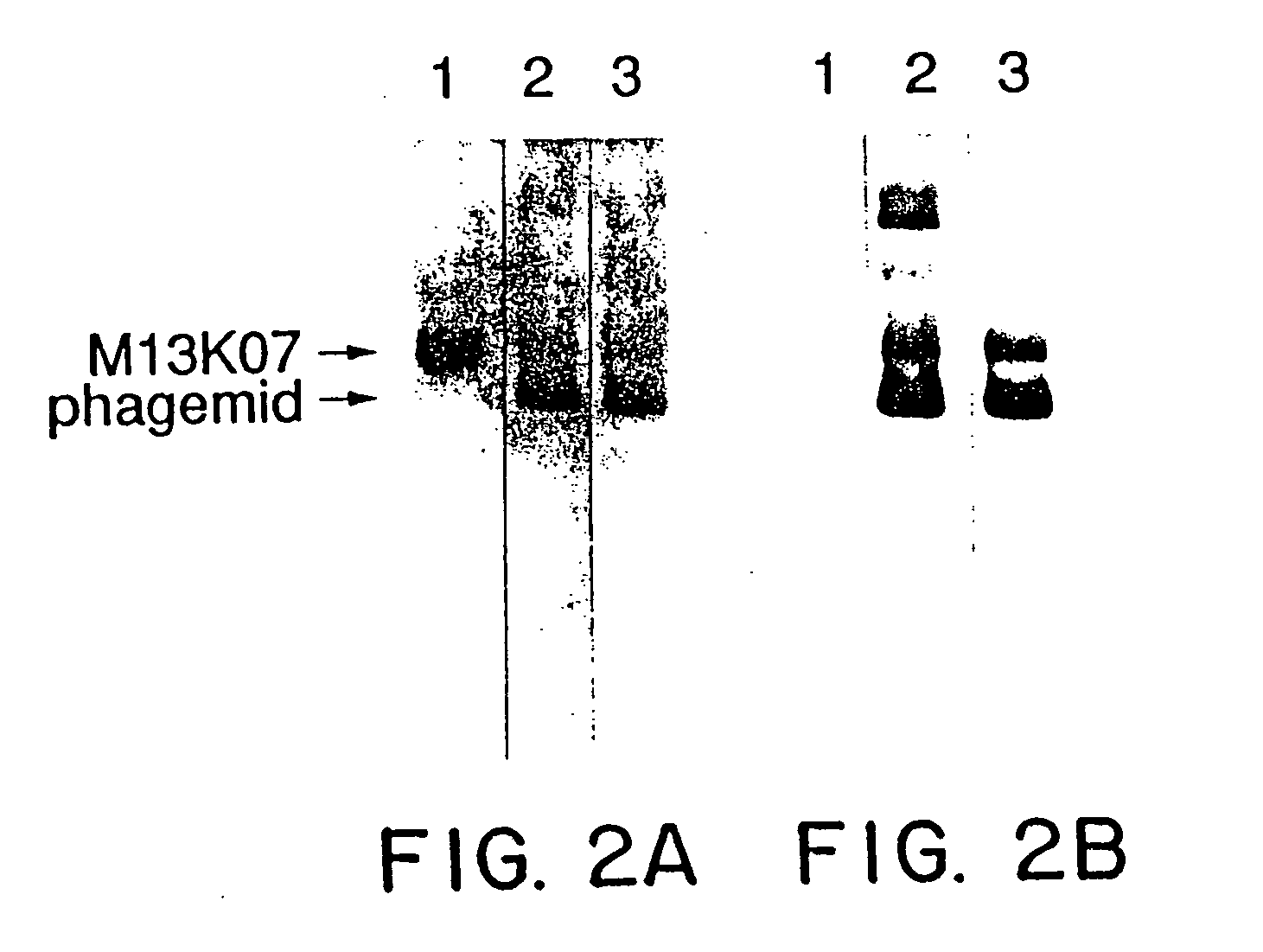
[0108] Rabbit polyclonal antibodies to hGH were purified with protein A, and coated onto microtiter plates (Nunc) at a concentration of 2 μg / ml in 50 mM sodium carbonate buffer (pH 10) at 4° C. for 16-20 hours. After washing in PBS containing 0.05% Tween 20, hGH or hGH-phagemid particles were serially diluted from 2.0-0.002 nM in buffer A (50 mM Tris (pH 7.5), 50 mM NaCl, 2 mM EDTA, 5 mg / ml bovine serum albumin, and 0.05% Tween 20). After 2 hours at room temperature (rt), the plates were washed well and the indicated Mab (Cunningham et al. supra) was added at 1 μg / ml in buffer A for 2 hours at rt. Following washing, horseradish peroxidase conjugated goat anti-mouse IgG antibody was bound at rt for 1 hour. After a final wash, the peroxidase activity was assayed with the substrate, o-phenylenediamine.
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| pH | aaaaa | aaaaa |
| temperatures | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com