One-step reduction and alkylation of proteins
a protein and alkylation technology, applied in the field of analysis of proteins, can solve the problems of time-consuming, opportunity to lose all or a portion of the protein sample, and achieve the effects of reducing the amount of time involved
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
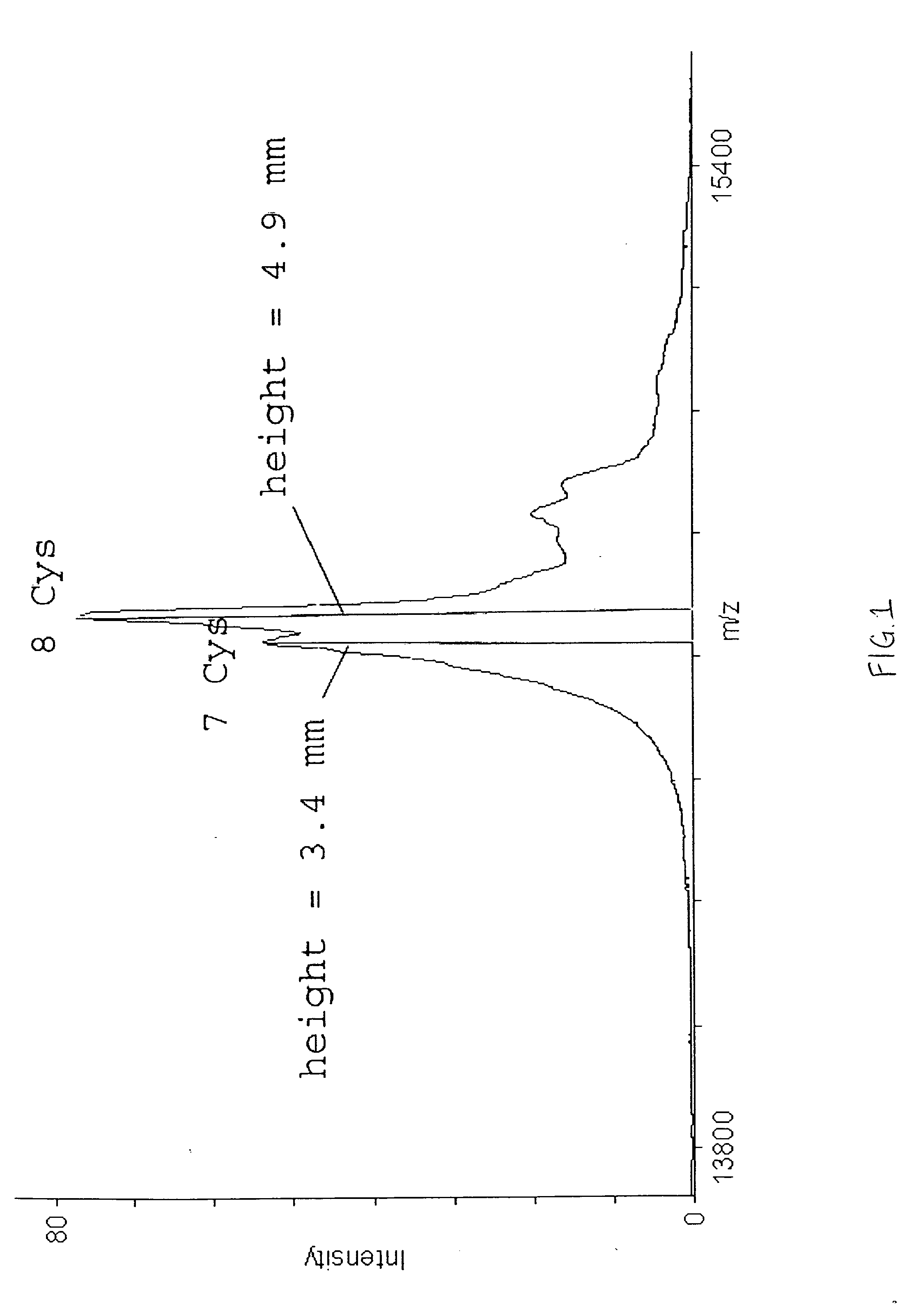
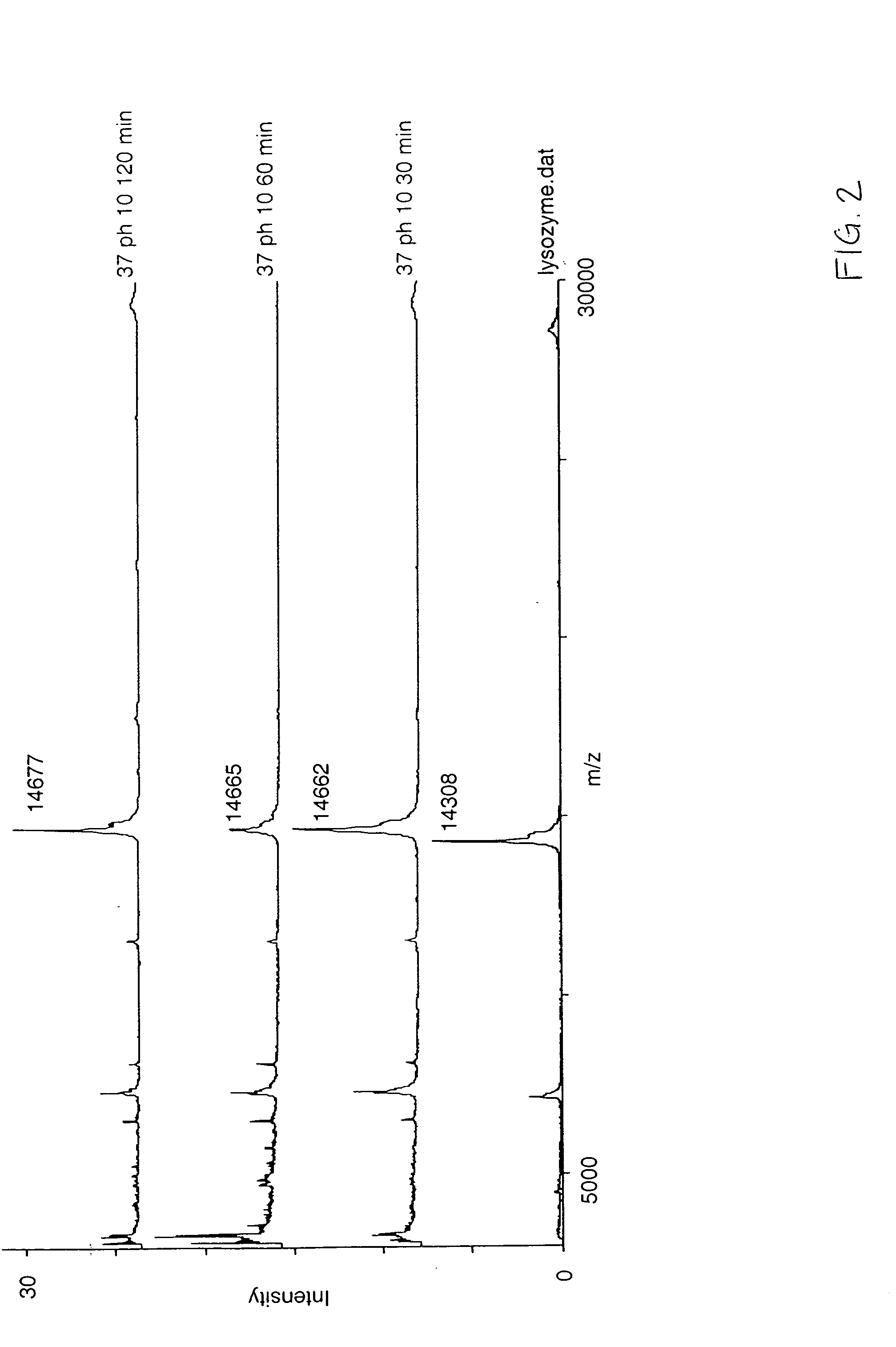
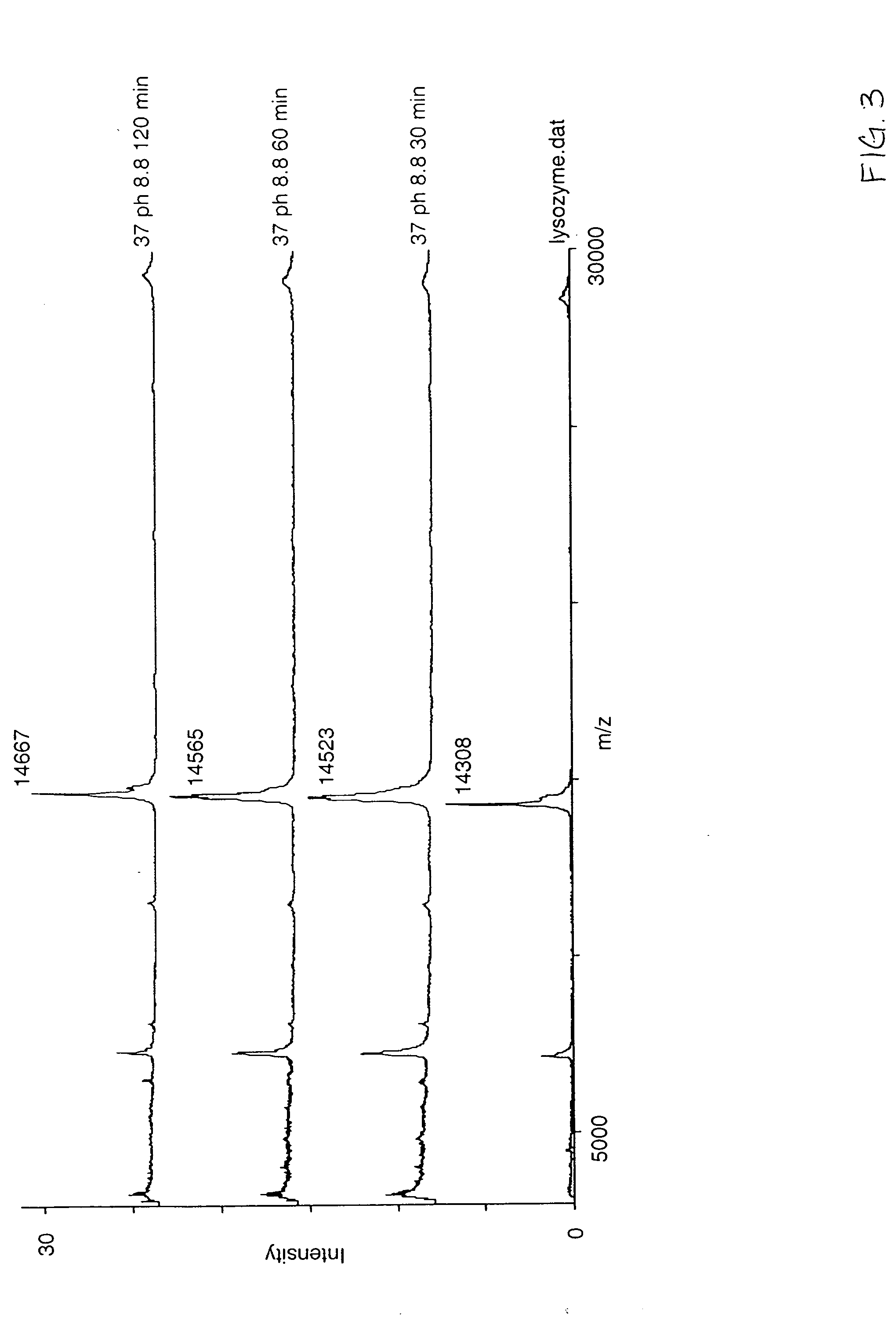
[0035] A purified lysozyme protein (having a mass of 14299.9 obtained by MALDI MS) in aqueous solution was adjusted to pH 10 with 0.1 M ammonium carbonate. An equal volume of the present reagent comprising 0.5% triethylphosphine, 2% iodoethanol and 97.5% acetonitrile was added to the protein solution. The protein / reagent mixture was capped and incubated at 37° C. for one hour and then uncapped and subjected to a vacuum for about an hour to produce a dried pellet in the container. The dried pellet was reconstituted in the same container to the original volume with a trypsin solution. The reconstituted protein was determined by MALDI MS to have a mass of 14650.9, a difference of 351, which corresponds to the addition of a 44 Da ethanol to each of the eight cysteines in the reduced and alkylated lysozyme.
example 2
[0036] The procedure of Example 1 was repeated with BSA substituted lysozyme. The BSA showed a gain in mass of 1539, which corresponds to a 44 Da alkylation for each of the 35 cysteines in BSA indicating the reduction and alkylation were successful.
example 3
[0037] An unfractionated serum sample was dried under vacuum. The resulting protein pellet was then redissolved in the same container with a volume of 8 M urea, and 0.1 M ammonium carbonate (pH 10) equal to the original volume of the serum sample in order to denature the proteins and maintain their solubility. An equal volume of the present reagent comprising 0.5% triethylphosphine, 2% iodoethanol and 97.5% acetonitrile was added to the protein solution. The protein / reagent mixture was capped and incubated at 37° C. for one hour and then uncapped and subjected to a vacuum for about an hour to produce a dried pellet in the container. The dried pellet was reconstituted in the same container with five times the original volume of a trypsin solution. The sites of alkylation were verified by subjecting the reconstituted material to LC / MS / MS analysis following incubation with trypsin. Sequence coverage as high as 90% has been observed by mass spectral analysis following trypsin digestion....
PUM
| Property | Measurement | Unit |
|---|---|---|
| pH | aaaaa | aaaaa |
| temperature | aaaaa | aaaaa |
| total peak height | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com