Selection of peptides with antibody-like properties
a technology of antibody-like properties and peptides, applied in the fields of protein chemistry, biochemistry, organic chemistry and molecular biology, can solve the problems of unfavorable high-throughput proteomic applications, and tedious and imperfect processes, and achieve the effect of facilitating binding of said complexes
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
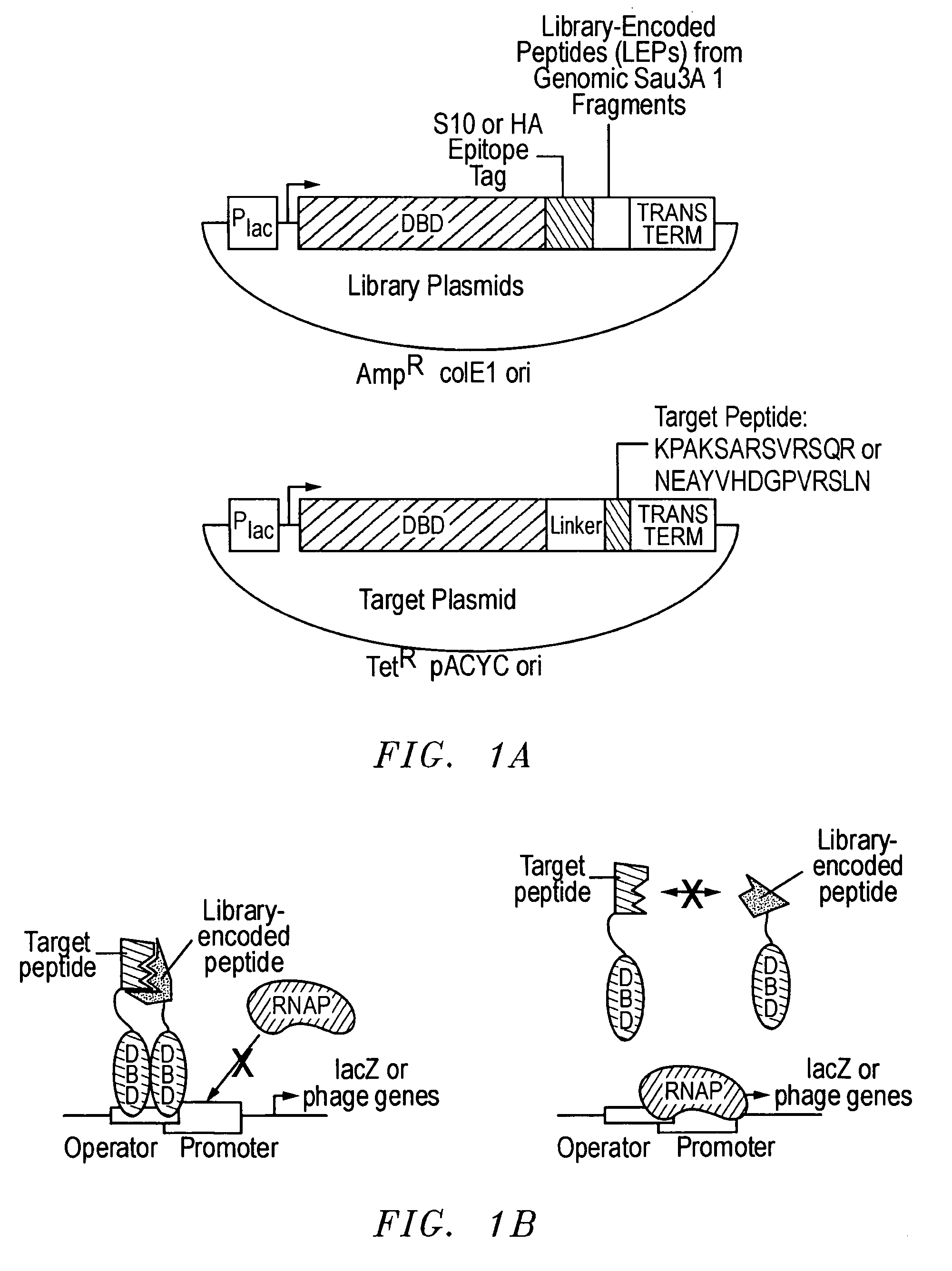
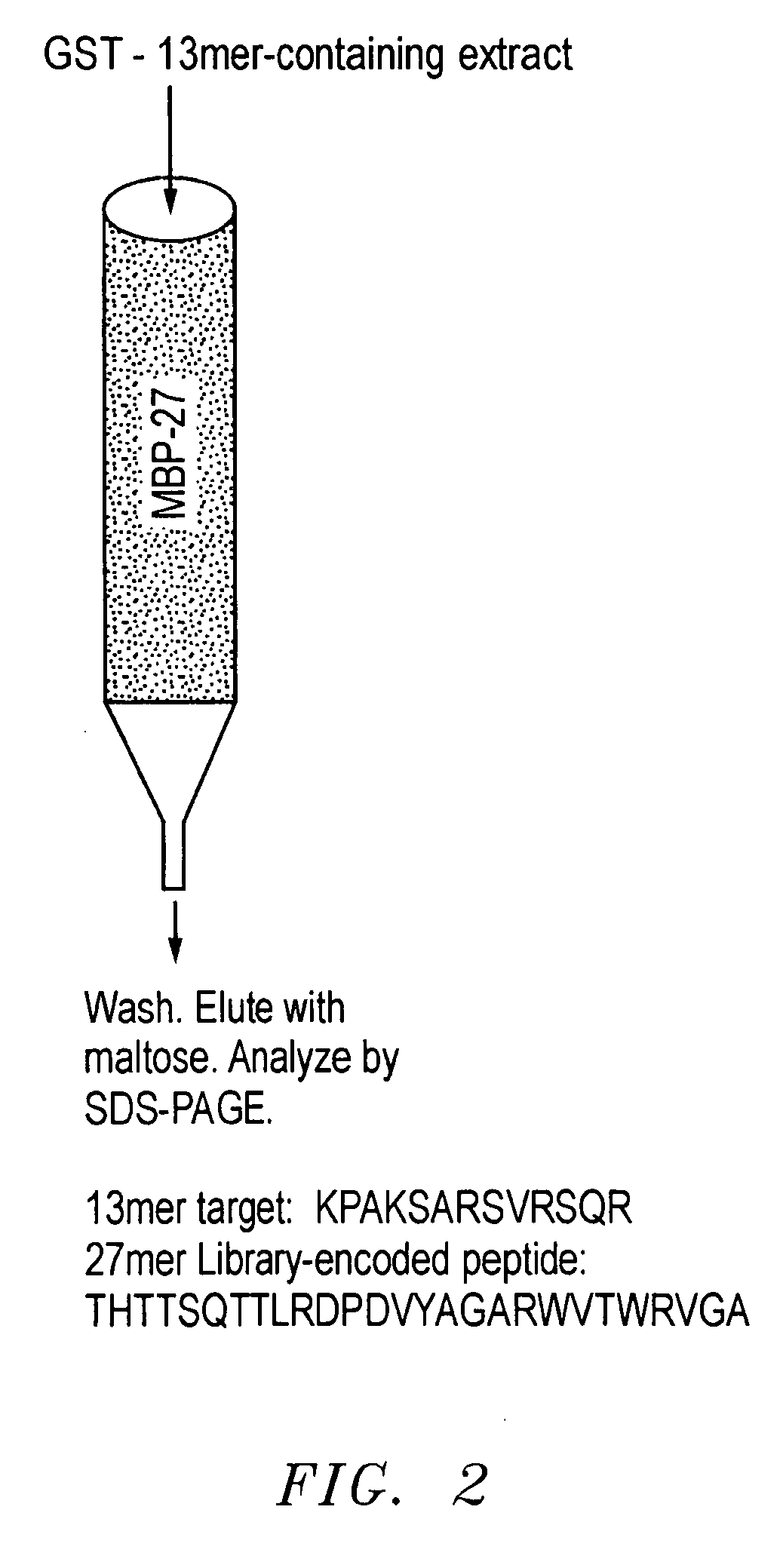
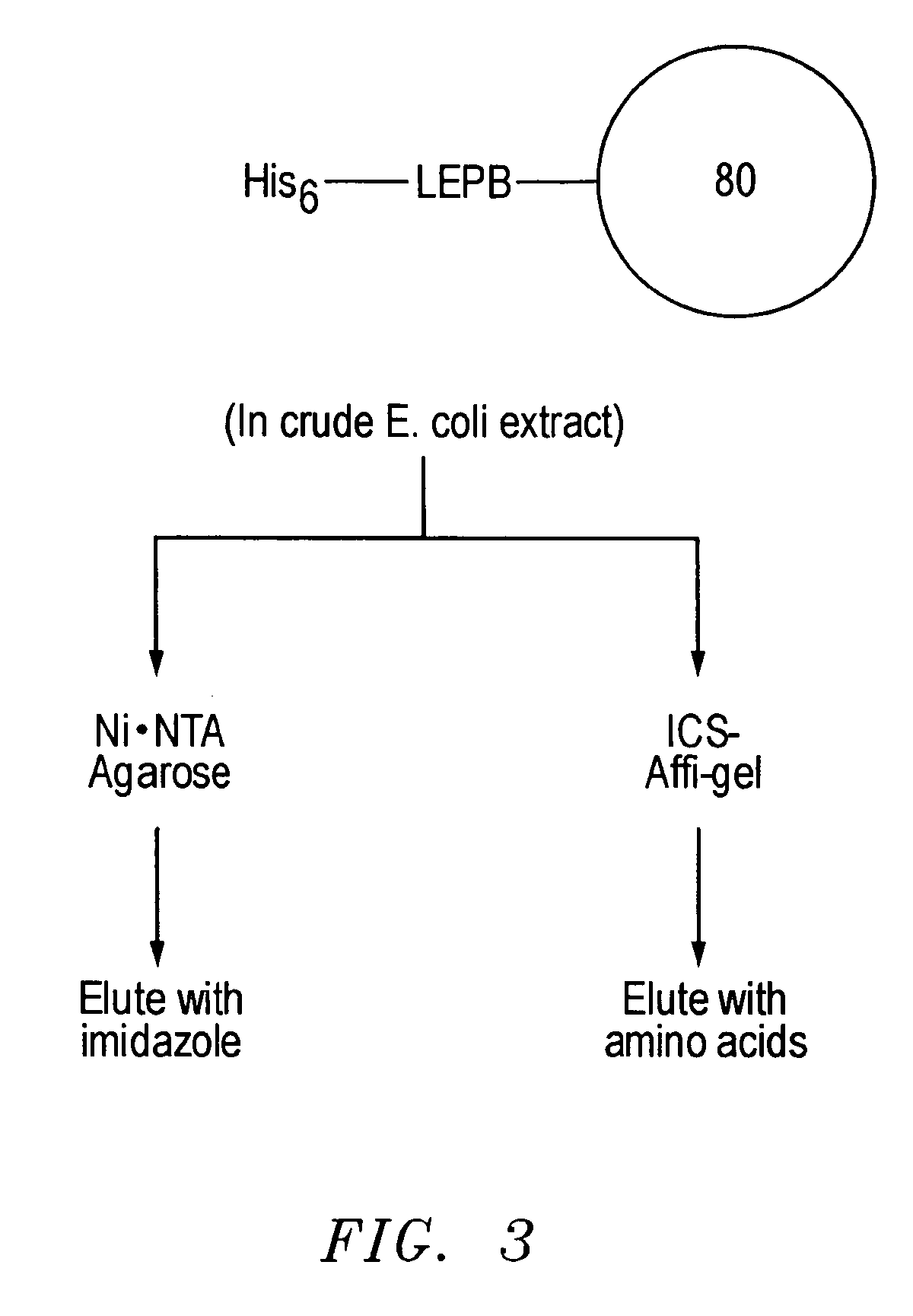
[0036] As discussed, an important goal in chemical biology is to be able to obtain specific ligands for any biomolecule of interest. Impressive advances have been made in isolating molecules, many of which are antibodies or antibody-derived, that bind proteins and nucleic acid targets with well-defined macromolecular structures. However, the identification of sequence-specific, peptide-binding ligands has been more difficult. In addition, even antibodies have major drawbacks: they are tedious and expensive to generate, difficult to produce in large quantities, are relatively fragile molecules unsuitable for certain field applications, and often bind so tightly that they or their target proteins are damaged upon attempted extraction. Natural peptide-binding proteins or protein domains have been mutagenized to derive species with novel binding specificities (Schneider et al., 1999), but like antibodies, these are globular macromolecules. Thus, though the development of synthetic recep...
PUM
| Property | Measurement | Unit |
|---|---|---|
| dissociation constant | aaaaa | aaaaa |
| dissociation constant | aaaaa | aaaaa |
| dissociation constant | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com