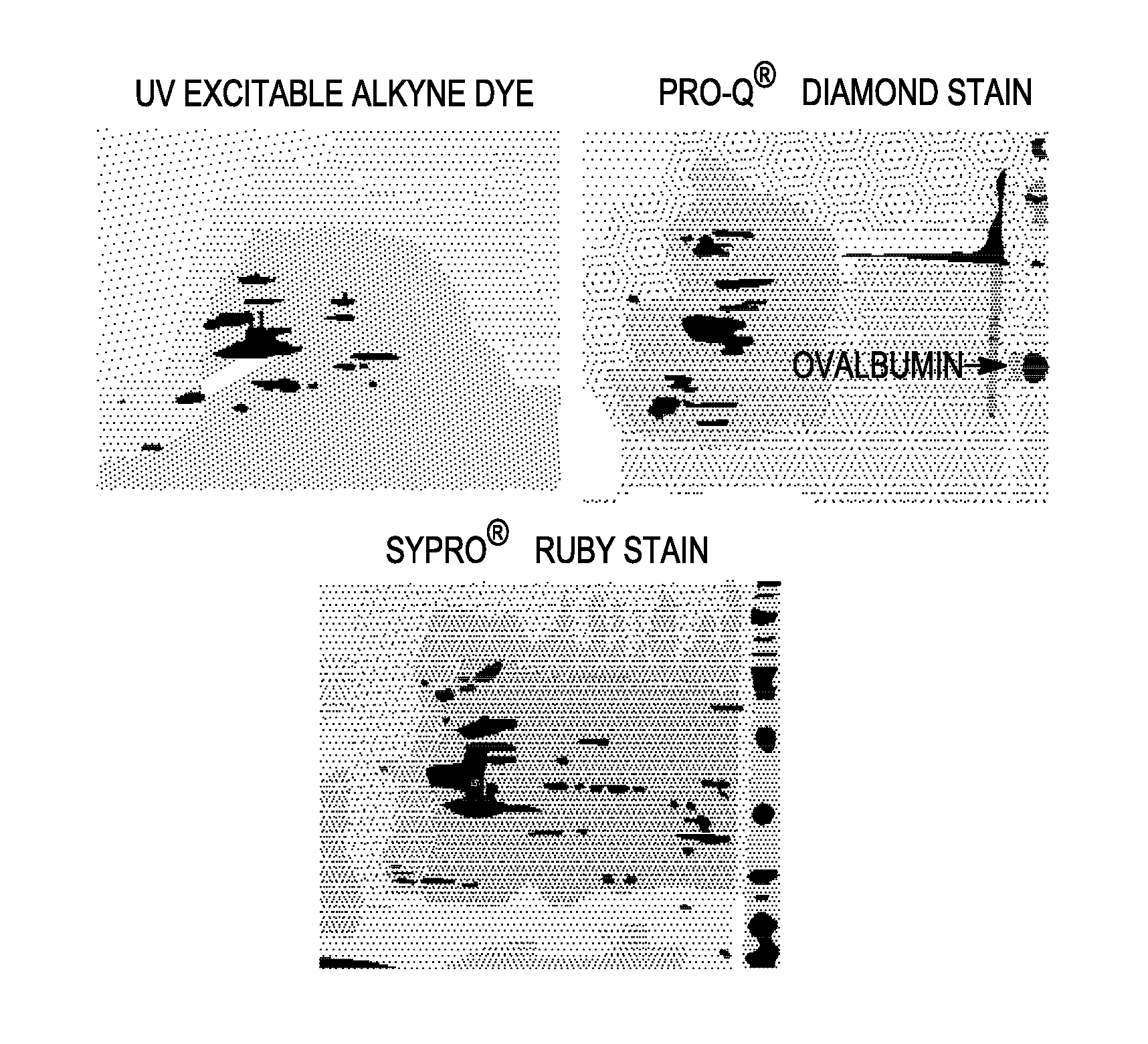
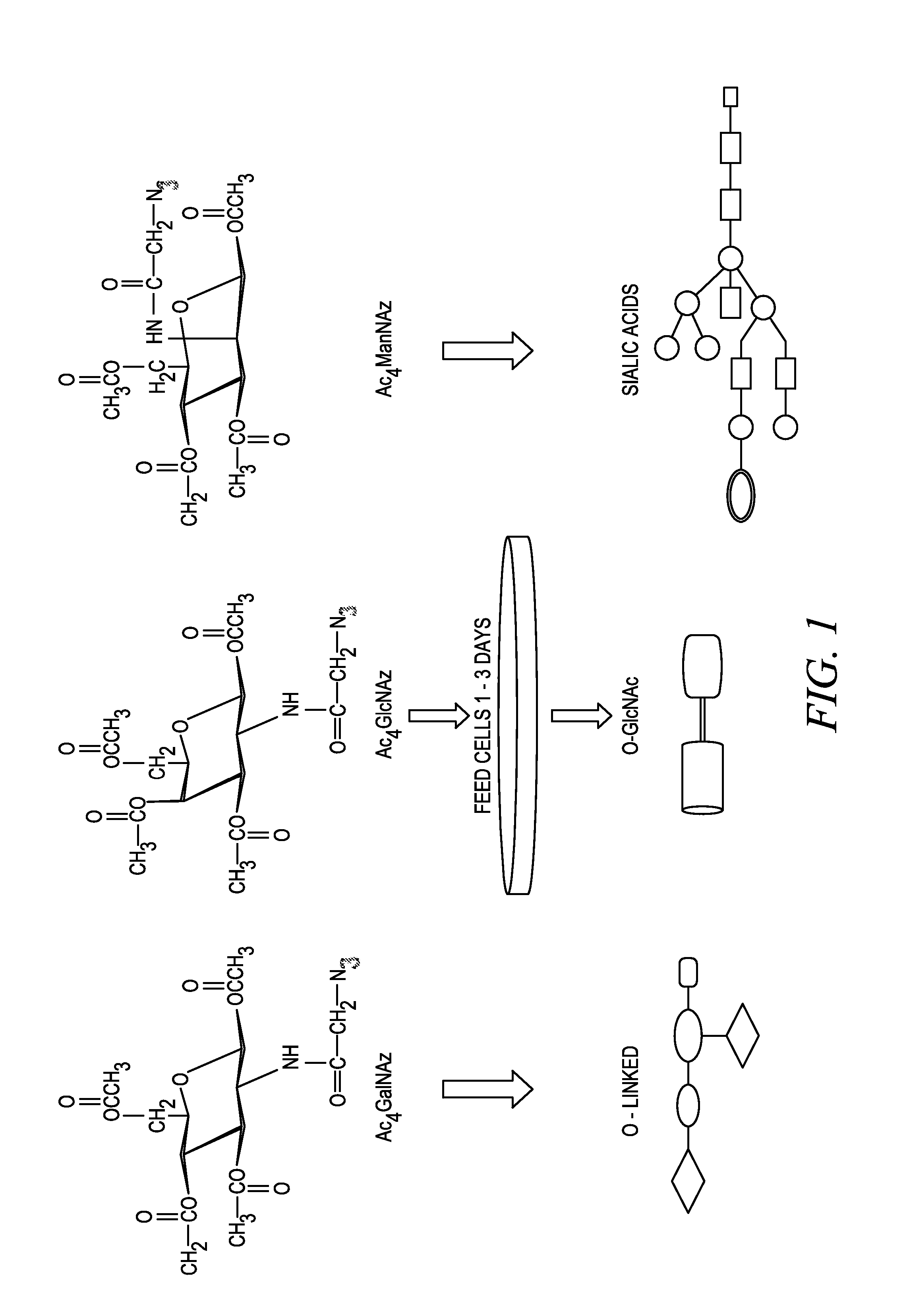
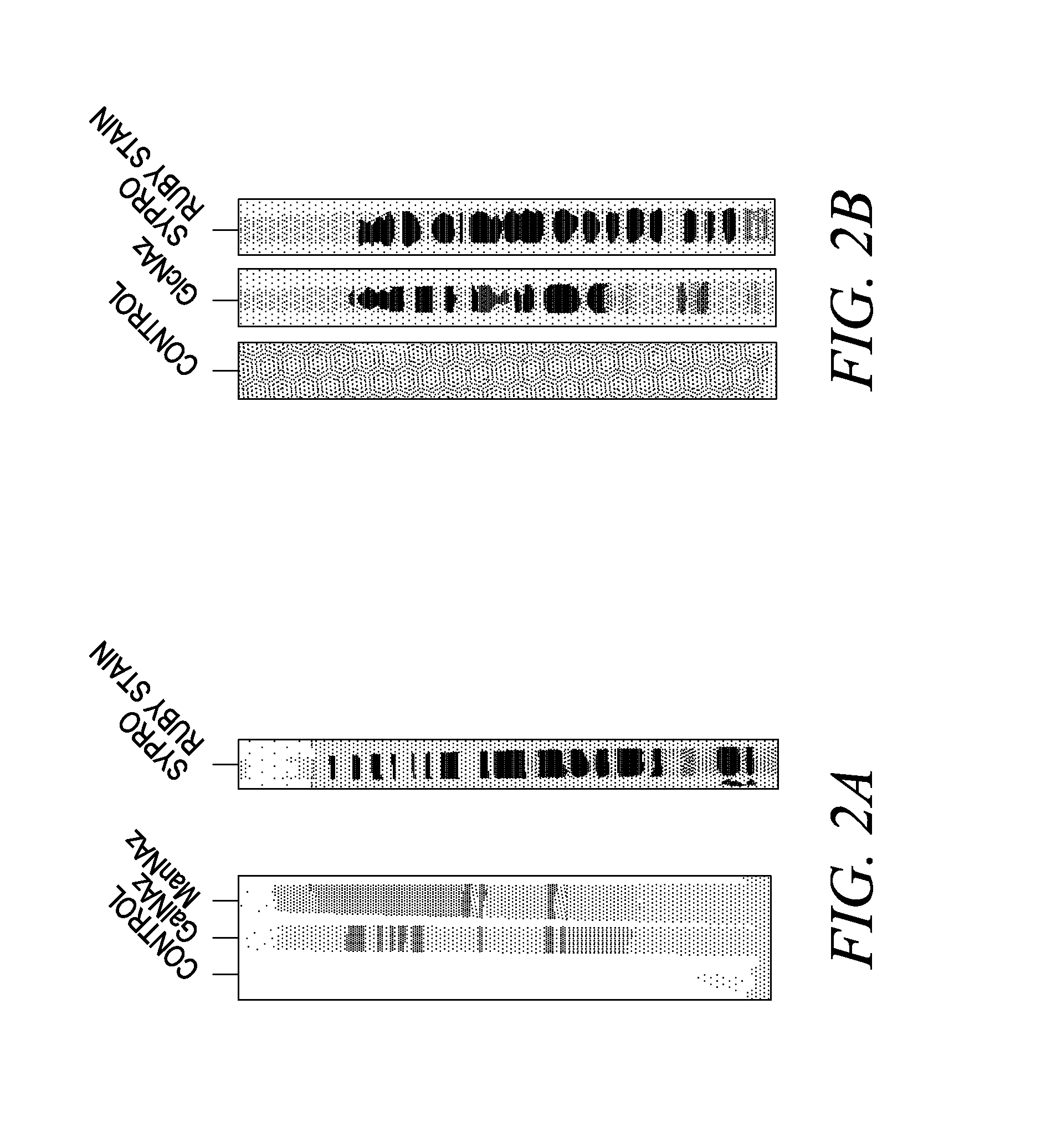
Labeling and detection of post translationally modified proteins
a technology of translationally modified proteins and labeling methods, applied in the field of labeling post translationally modified biomolecules, can solve the problems of inability to apply in-gel fluorescence detection of modified proteins, and the method is devoid of the typical problems encountered in antibody detection on blots
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Synthesis of UDPGalNAz
[0269] The synthesis of UDPGalNAz is shown in the following reaction scheme.
[0270] Azidoacetic acid. To a solution of iodoacetic acid (8.0 g, 43.0 mmol) and H2O (100 mL) was added sodium azide (5.62 g, 86.0 mmol). The solution was stirred at RT and protected from light. After 4 days, the solution was diluted with 1 N HCl (30 mL) and the pH was check to ensure that it was in the range of 2-3. The solution was extracted with EtOAc (2×100 mL), then the combined organics were washed with saturated NaHSO3 (1×50 mL), brine (1×50 mL) and dried over MgSO4. The solution was decanted, concentrated and the crude azidoacetic acid (3.34 g, 53%) was used directly in the next step without further purification.
[0271] N-Azidoacetylgalactosamine (mix of anomers). To a solution of azidoacetic acid (3.34 g, 33.06 mmol) in methanol (170 mL) was added D-galactosamine hydrochloride (5.09 g, 23.62 mmol) followed by triethylamine (7.90 mL, 56.68 mmol). This solution was stirred at ...
example 2
Synthesis of UMP-N-methylimidazolide
[0276] The synthesis of UMP-N-methylimidazolide is shown in the following reaction scheme.
[0277] Uridine 5′-monophosphate triethylammonium salt (5′-UMP triethylammonium salt). Uridine 5′-monophosphate disodium salt (1.0 g) was dissolved in H2O (2 ml) and passed through Dowex resin triethylammonium salt (2.7 cm×7.0 cm column size) with H2O as eluent. All of the eluent was concentrated and after the UV active material ceased eluting from the column. The solution was concentrated and co-evaporated with toluene to until a white, crystalline solid was obtained (0.78 g, 80%).
[0278] Uridine monophosphate-N-methylimidazole (UMP-N-methylimidazole). To a suspension of uridine 5′-monophosphate triethylammonium salt (0.097 g, 0.22 mmol) in CH3CN (0.70 mL) was added N,N-dimethylaniline (0.11 mL, 0.864 mmol), and triethylamine (0.03 mL, 0.22 mmol). The suspension was cooled to 0° C. In a separate flask, trifluoroacetic anhydride (0.15 mL, 1.08 mmol) was add...
example 3
Synthesis of Dapoxyl® alkyne
[0279] The synthesis of Dapoxyl® alkyne is shown in the following reaction scheme.
[0280] To a solution of Dapoxyl® carboxylic acid, succinimidyl ester (50 mg, 0.12 mmol) in DMF (0.4 mL) at RT was added propargylamine (42 μL, 0.61 mmol). The initial clear orange solution turned yellow and cloudy. After ˜15 min at RT the reaction was complete, and the solution was concentrated to dryness. The residue was purified via HPLC to afford the product (36 mg, 84%). TLC (10% EtOAc, CHCl3) Rf=0.30; ESI m / z 346 (M+, C21H19N3O2 requires 346).
PUM
| Property | Measurement | Unit |
|---|---|---|
| mass | aaaaa | aaaaa |
| mass | aaaaa | aaaaa |
| mass | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com