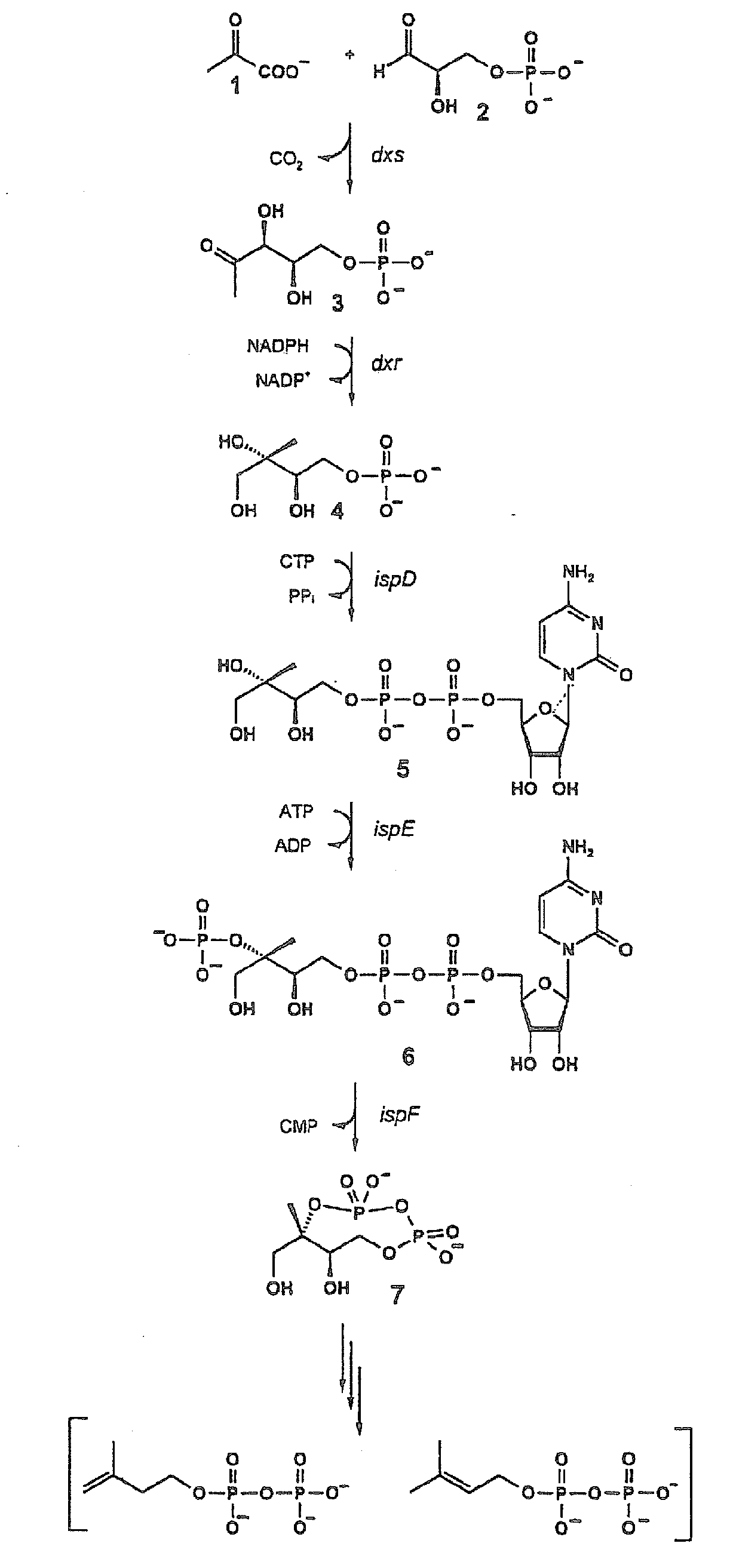
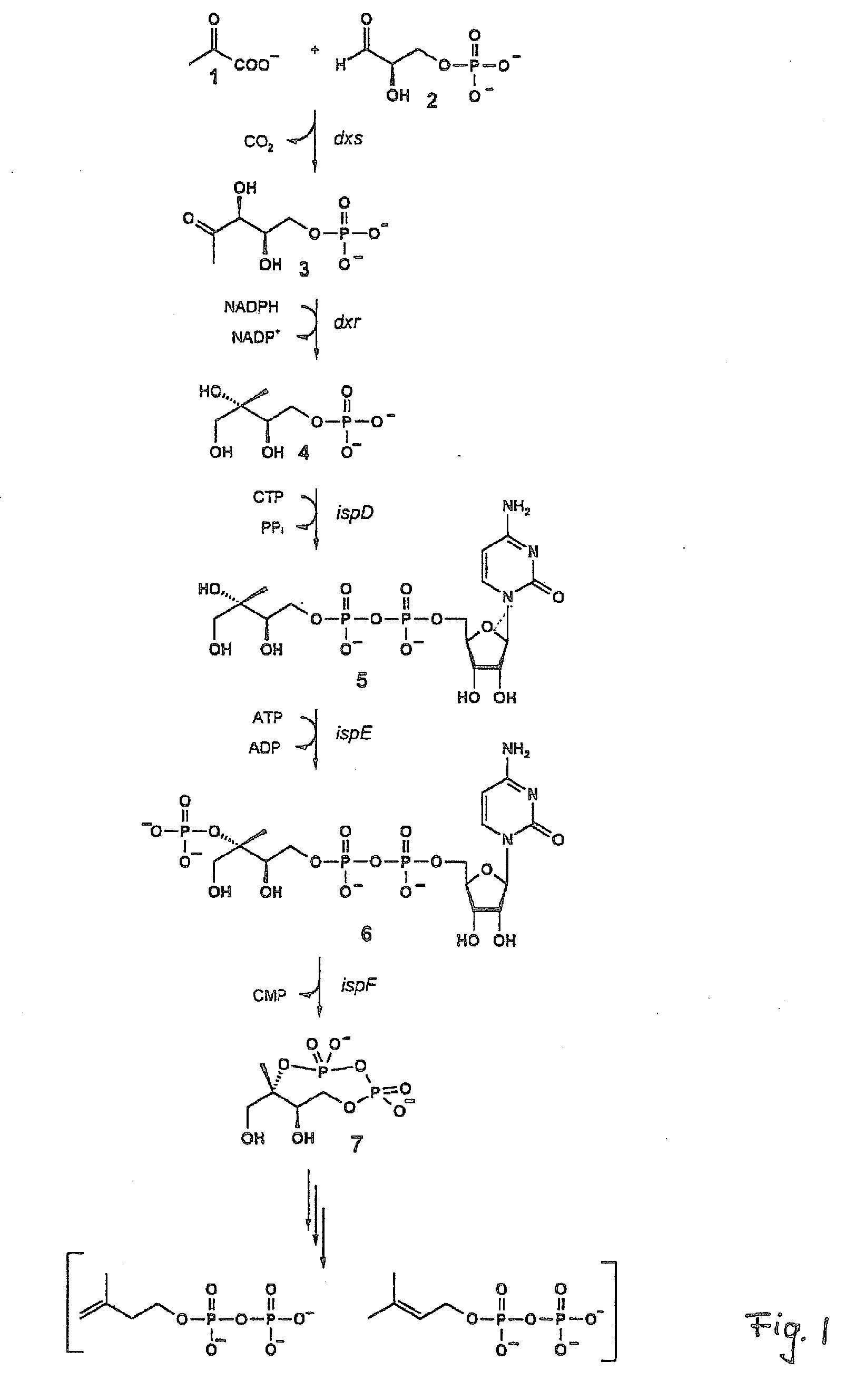
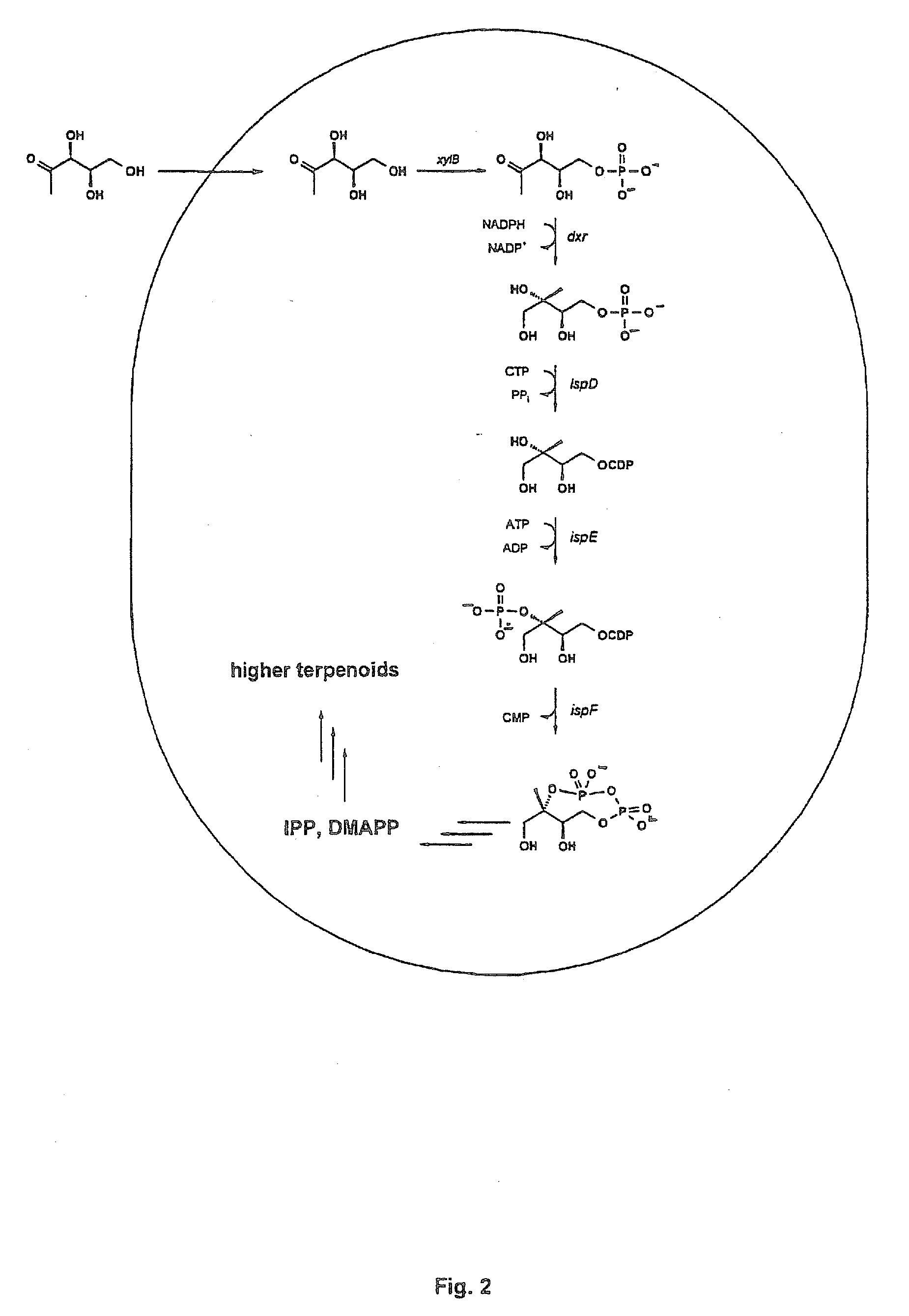
Intermediates and enzymes of the non-mevalonate isoprenoid pathway
a technology of intermediates and enzymes, which is applied in the direction of lyases, transferases, protozoa, etc., can solve the problems of hampered previous attempts to approach these goals and low biosynthesis rate along these pathways
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Construction of a Vector Carrying the xylB Gene of Escherichia coli Capable for Transcription and Expression of D-xylulokinase
[0147]Chromosomal DNA from Escherichia coli strain XL1-Blue (Bullock et al. 1987; commercial source: Stratagene, LaJolla, Calif., USA) is isolated according to a method described by Meade et al. 1982.
[0148]The E. coli ORF xylB (accession no. gb AE000433) from base pair (bp) position 8596 to 10144 is amplified by PCR using chromosomal E. coli DNA as template. The reaction mixture contains 10 pmol of the primer 5′-CCGTCGGAATTCGAGGAGAAATTAACCATGTATATCGGGATAGATCTTGG-3′ (SEQ ID NO:1), 10 pmol of the primer 5′-GCAGTGAAGCTTTTACGCCATTAATGGCAGAAGTTGC-3′ (SEQ ID NO:2), 20 ng of chromosomal DNA, 2 U of Taq DNA polymerase (Eurogentec, Seraing, Belgium) and 20 nmol of dNTPs in a total volume of 100 μl containing 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-hydrochloride, pH 8.8 and 0.1% (w / w) Triton X-100.
[0149]The mixture is denaturated for 3 min at 94° C. Then 30 PCR cycles for ...
example 2
Construction of a Vector Carrying the XylB and dxr Genes of Escherichia coli Capable for Transcription and Expression of D-xylulokinase and DXP reductoisomerase
[0154]The E. coli ORF dxr (accession no. gb AE000126) from base pair (bp) position 9887 to 11083 is amplified by PCR using chromosomal E. coli DNA as template. The reaction mixture contains 10 pmol of the primer 5′-CTAGCCAAGCTTGAGGAGAAATTAACCATGAAGCAACTCACCATTCTGG-3′ (SEQ ID NO:3), 10 pmol of the primer 5′-GGAGATGTCGACTCAGCTTGCGAGACGC-3′ (SEQ ID NO:4), 20 ng of chromosomal DNA, 2 U of Taq DNA polymerase (Eurogentec) and 20 nmol of dNTPs in a total volume of 100 μl containing 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-hydrochloride, pH 8.8 and 0.1% (w / w) Triton X-100.
[0155]The mixture is denaturated for 3 min at 94° C. Then 30 PCR cycles for 60 sec at 94° C., 60 sec at 50° C. and 75 sec at 72° C. followed. After further incubation for 10 min at 72° C., the mixture is cooled to 4° C. An aliquot of 2 μl is subjected to agarose gel elec...
example 3
Construction of a Vector Carrying the XylB, dxr and ispD Genes of Escherichia coli Capable for Transcription and Expression of D-xylulokinase, DXP Reductoisomerase and CDP-ME Synthase
[0161]The E. coli ORF ispD (accession no. gb AE000358) from base pair (bp) position 6754 to 7464 is amplified by PCR using chromosomal E. coli DNA as template. The reaction mixture contains 10 pmol of the primer 5′-CCGGGAGTCGACGAGGAGAAATTAACCATGGCAACCACTCATTTGGATG-3′ (SEQ ID NO:5), 10 pmol of the primer 5′-GTCCAACTCGAGTTATGTATTCTCCTTGATGG-3′ (SEQ ID NO:6), 20 ng of chromosomal DNA, 2 U of Taq DNA polymerase (Eurogentec) and 20 nmol of dNTPs in a total volume of 100 μl containing 1.5 mM MgCl2, 50 mM KCl, 10 mM Tris-hydrochloride, pH 8.8 and 0.1% (w / w) Triton X-100.
[0162]The mixture is denaturated for 3 min at 94° C. Then 30 PCR cycles for 30 sec at 94° C., 30 sec at 50° C. and 45 sec at 72° C. followed. After further incubation for 10 min at 72° C., the mixture is cooled to 4° C. An aliquot of 2 μl is su...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com