Method for detecting nanbv associated seroconversion
a seroconversion and nanbv technology, applied in the field of recombinant expression vectors, can solve the problems of increasing false positives, limited development of antibodies detection immunoassays, and reducing assay specificity and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Isolation of the HIV p24 Gene and Construction of Expression Vector
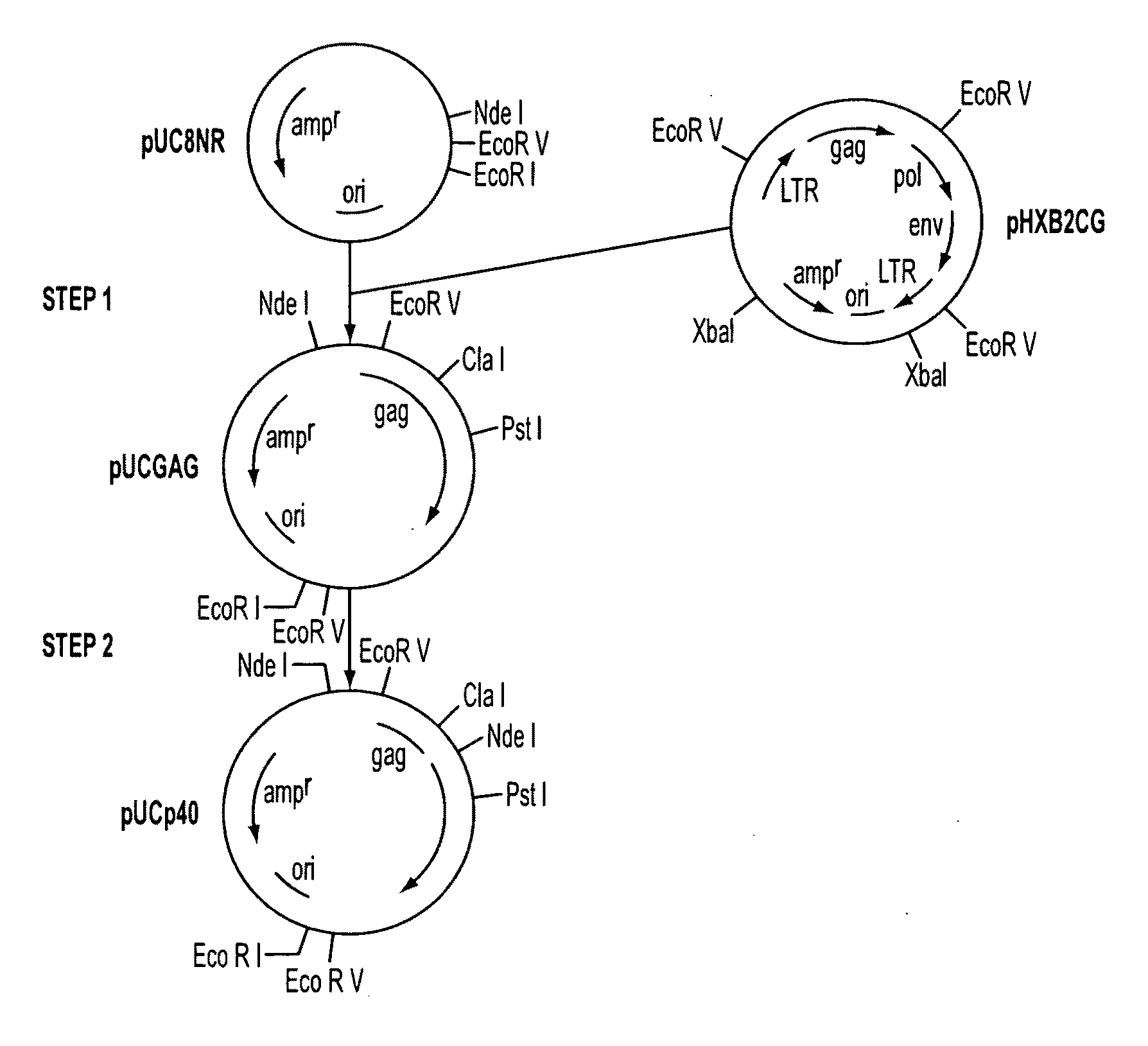
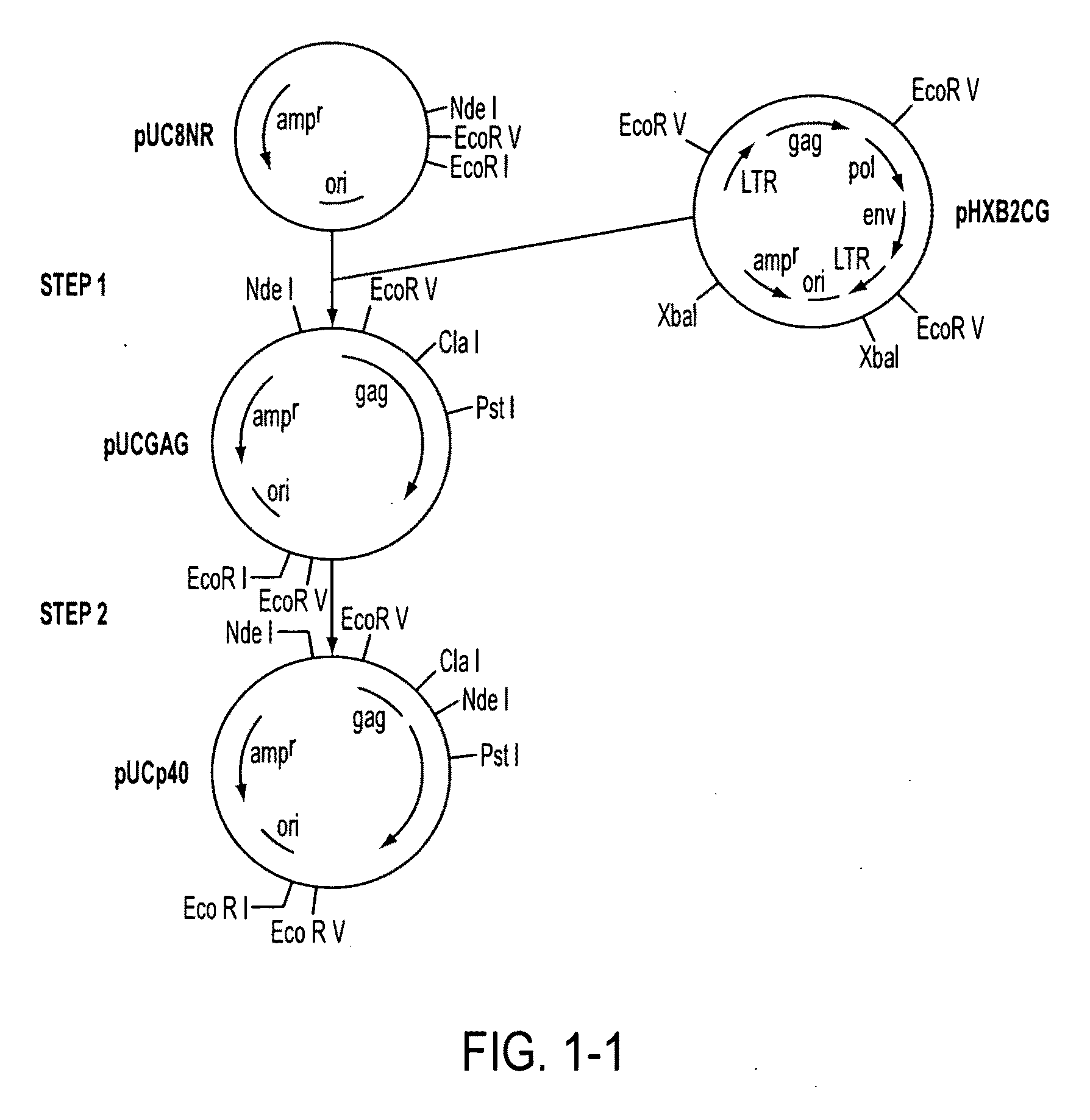
[0110]The gag region from the pHXB2CG plasmid clone of HTLV IIIB (obtained from Dr. Robert Gallo, National Cancer Institute, Bethesda, Md.) was isolated by EcoRV restriction enzyme digestion of plasmid pHXB2CG and the resulting 2.86 kilobase fragment was isolated and inserted by ligation into the EcoRV site of a modified pUC8 vector (pUC8NR) to form plasmid PUCGAG (FIG. 1, Step 1).
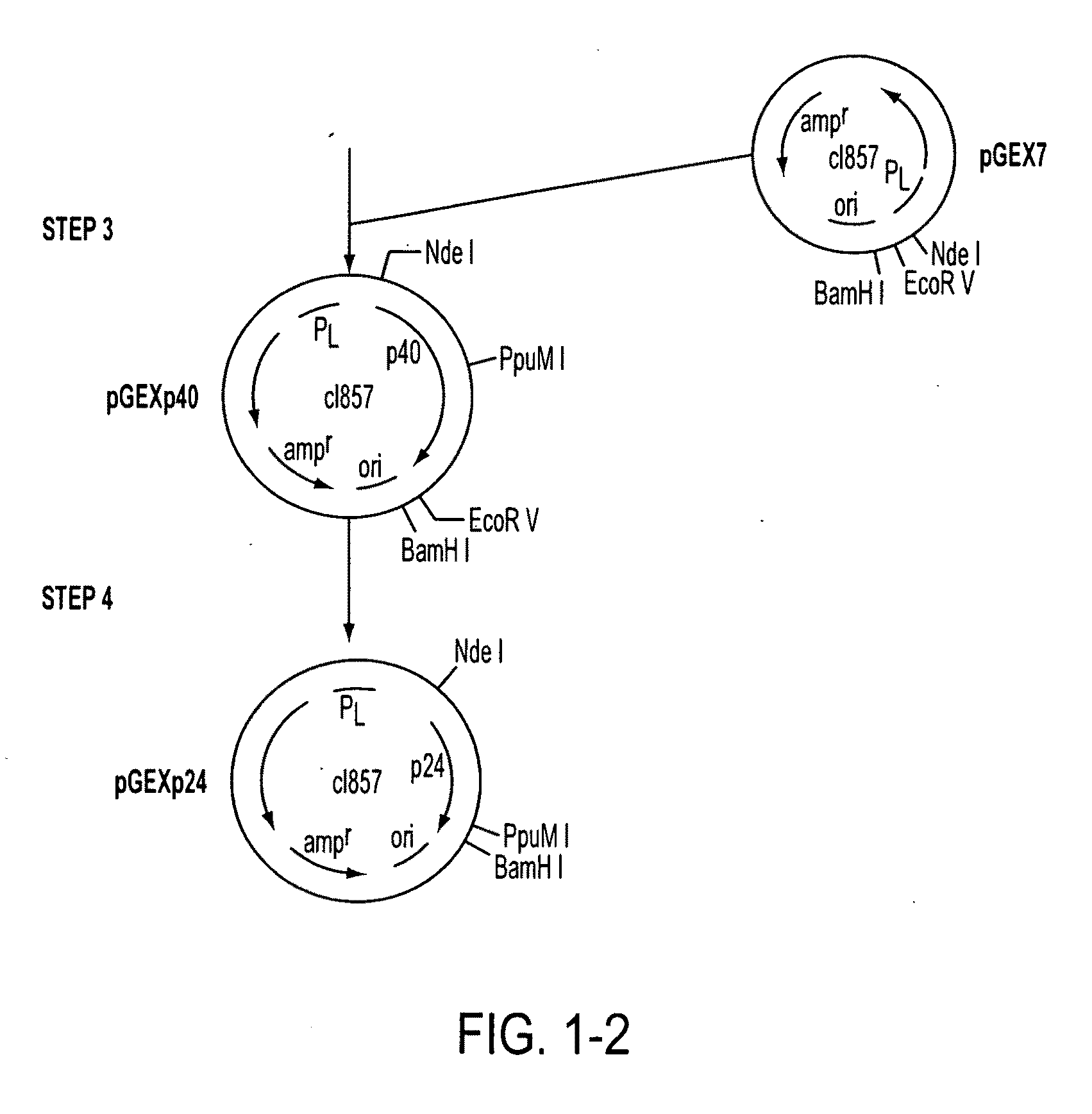
[0111]The plasmid (PUCGAG) was mutagenized to generate an ATG translational initiation codon and an NdeI restriction enzyme site (CATATG) at the beginning of the p24 structural gene by the following series of manipulations (FIG. 1, Step 2). After transformation of pUCGAG into the methylation deficient dam-strain of E. coli, New England Biolabs, a gap was created in the pUCGAG DNA at the p24 amino terminus by cutting with the ClaI and PstI restriction enzymes to form gapped pUCGAG that lacks the smaller DNA segment from the p24 amino terminu...
example 2
Formation of Composite DNAs Comprising the pGEXp24 Vector with an Inserted Gene for a Conserved Envelope gp41 (Subtype 0) Antigen
[0120]The plasmid pGEXp24, was linearized by digestion with the restriction enzyme PpuMI and purified by phenol-chloroform extraction followed by precipitation with ethanol. Two complementary oligonucleotides (sequences given by nucleotides 686 to 763 and the complement of nucleotides 689 to 766 of SEQ ID NO:1) forming protruding cohesive termini when annealed, were synthesized. The synthetic oligonucleotides were allowed to form a duplex by mixing and heating to 90° C. for a approximately 3 minutes, followed by annealing at room temperature for a period of 10 minutes. The hybrid molecule represents a hybrid gene sequence encoding the p24 molecule interrupted after codon 225 by a linker amino acid (lysine), envelope sequence (amino acids 227-249) for the conserved region of HIV Subtype 0 gp41 polypeptide, strain ANT, followed by a repetition of p24 residue...
example 3
Purification of Recombinant p24-gp41 (subtype 0) Fusion Proteins
[0124]Plasmids containing the lambda promoter (pL) are normally carried in a strain of bacteria containing a lysogen of bacteriophage lambda in order to minimize the expression of the gene product of interest during the manipulation of DNAs. The pGEX7-based plasmids described in Example 1 were all carried in a lysogen of the MM294 strain of E. coli. Expression from the lambda promoter of pGEX7 can be demonstrated by transfer of the plasmid into an uninfected bacterial host (e.g., E. coli strain W3110, accession no. #27325, ATCC, Rockville, Md.) and inactivation of the cl repressor protein at 42° C. Competent E. coli (strain W3110, 100 μl bacterial suspension) were transformed with 1 μl of pGEXp24gp41-ANT, pGEXp24gp41-MVP or pGEXp24gp41-X84328. After 60 minutes on ice, the bacteria were diluted to 1 ml with LB medium and incubated for a further 60 minutes at 30° C. Aliquots of the culture were than plated on ampicillin c...
PUM
| Property | Measurement | Unit |
|---|---|---|
| apparent relative molecular weight | aaaaa | aaaaa |
| diameter | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com