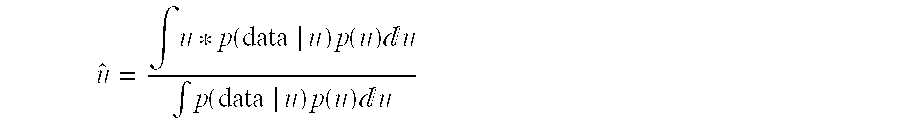Artificial selection method and reagents
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Model Artificial Selection Method for a Holstein Cattle Population
Rationale
[0289]Many breeds of livestock have a small effective population size of 50-100, including the Holstein cattle population. This means that most chromosome segments found in animals of the current generation trace back to one of less than about 100 key ancestors within a few generations. This short coalescence time means that the chromosome segments are large and could be recognised by their haplotype at a group of markers. Consequently we can carry out ‘in silico’ genotyping as follows:[0290]1. Genotyping key ancestors for a dense set of markers;[0291]2. Genotyping individuals of the current population / generation for sufficient markers to permit chromosome segments to be matched to the segments carried by the key ancestors; and[0292]3. Inferring genotypes of individuals in the current population / generation to be the same as those of the key ancestor or founder for the matching chromosome segment.
[0293]By this...
PUM
| Property | Measurement | Unit |
|---|---|---|
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
| Volume | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com