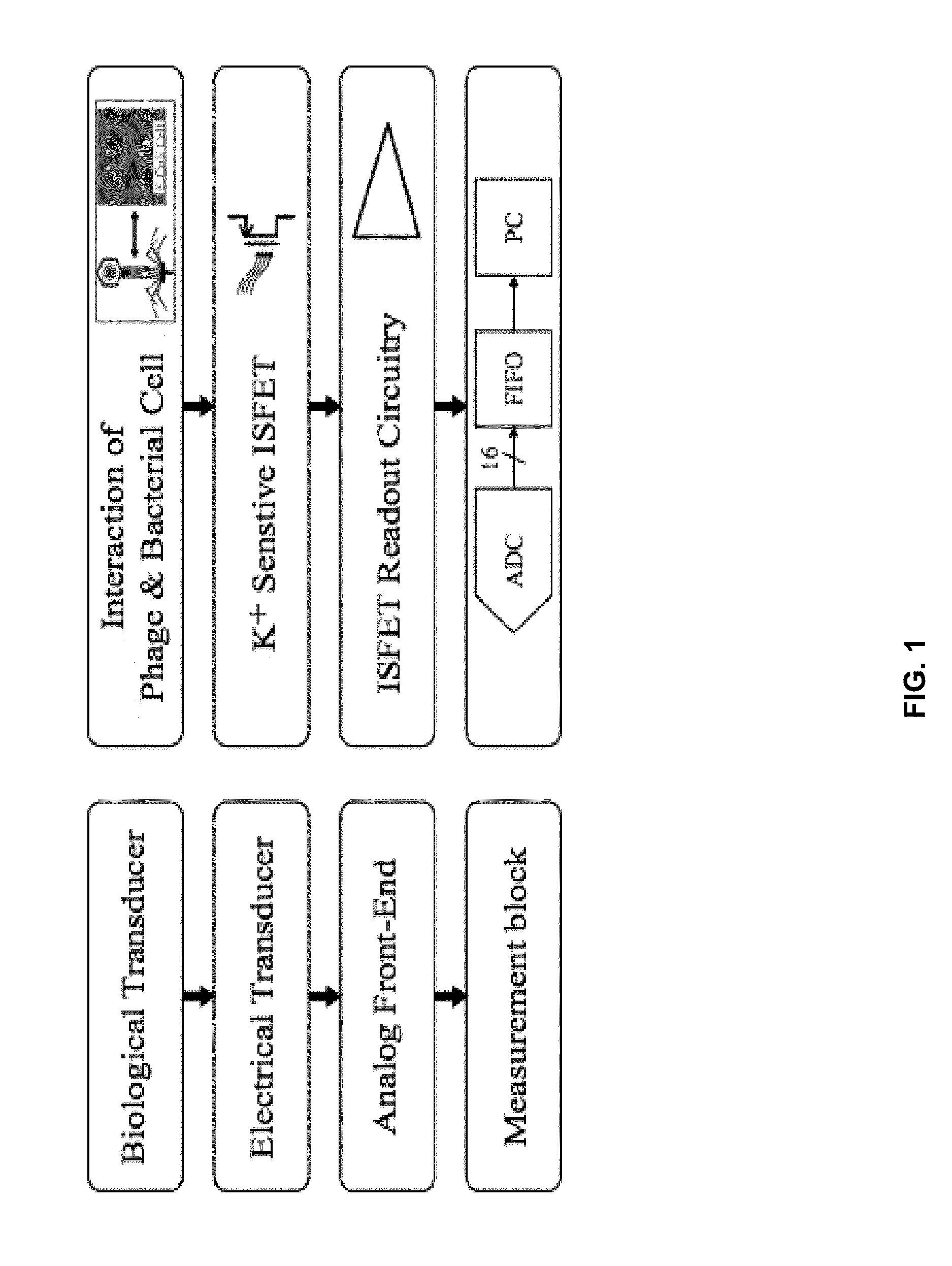
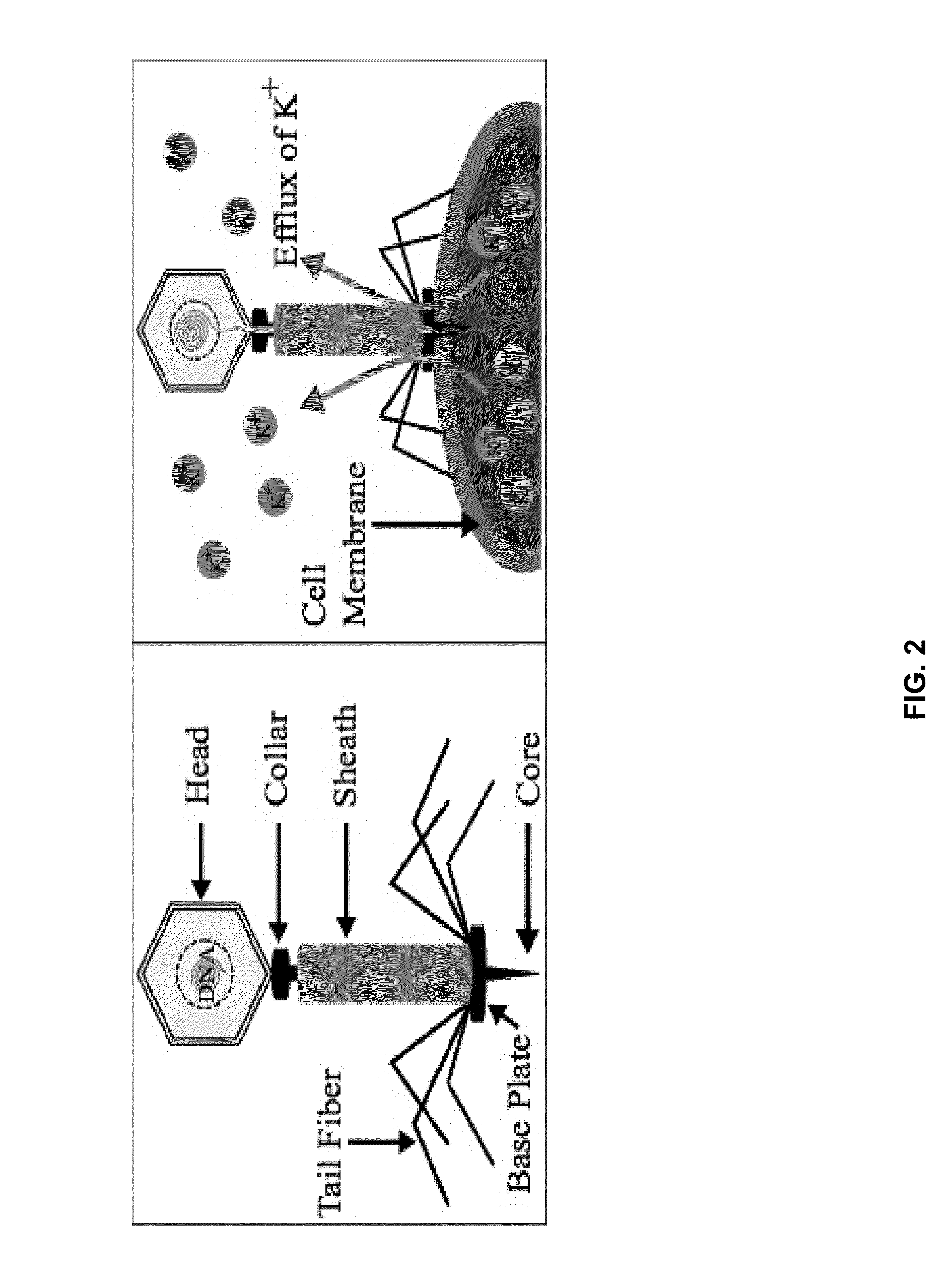
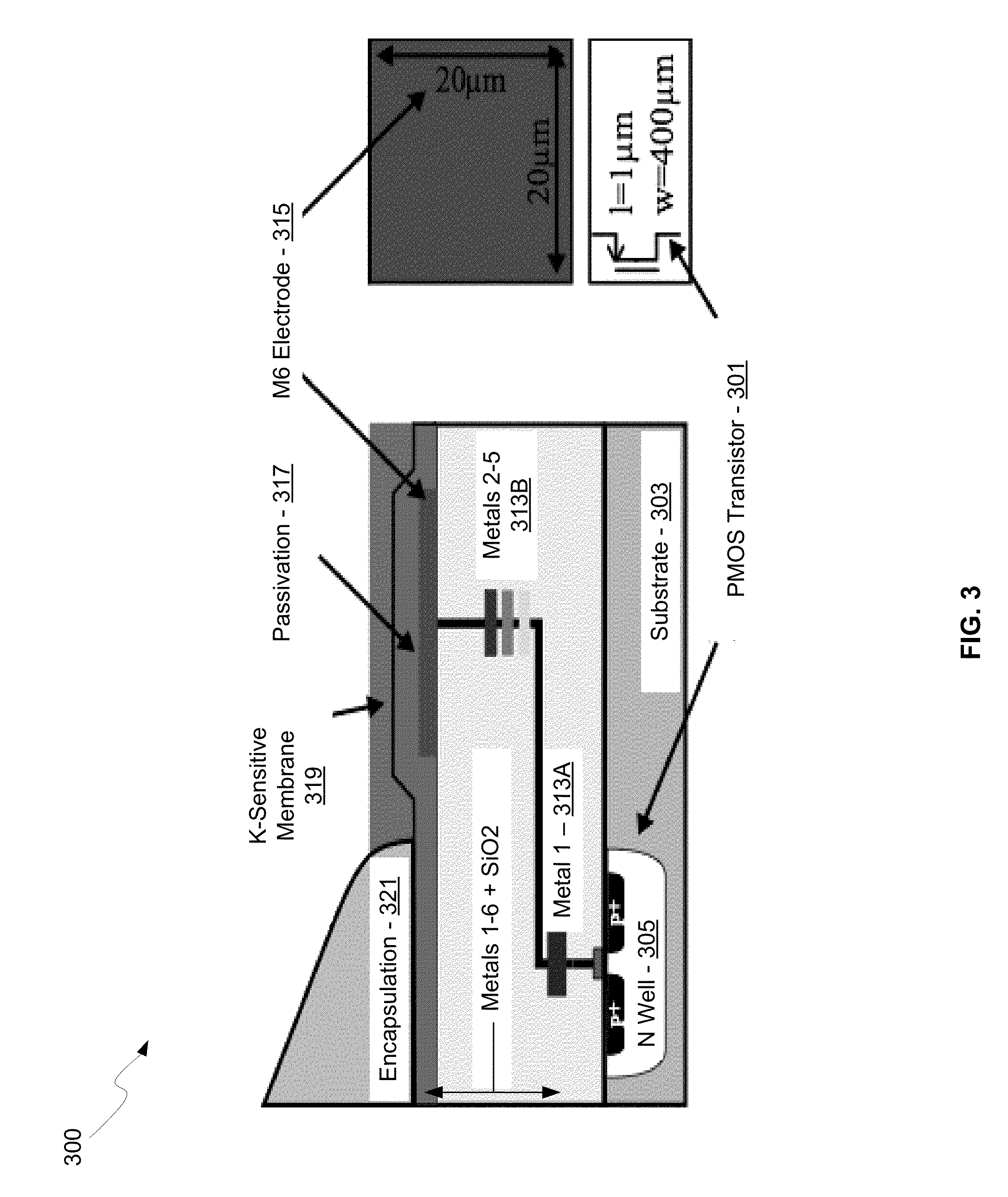
Integrated sensor for the rapid identification of bacteria using isfets
a sensor and bacteria technology, applied in the field of integrated sensors for the rapid identification of bacteria using isfets, can solve the problems of laborious sample preparation, difficult sample preparation, and difficult preparation of samples, and achieve the effects of reducing the number of samples
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
[0056]Live bacterial cells and phages were used on the chip in demonstration of the ISFET system 600. Before starting the experiments, the chips may be cleaned and potassium-sensitive ISFET chips prepared as explained in the previous sections.
[0057]The measurements were done using two strains of like E.coli K-12 bacterial cells. The positive control experiments were performed using E.coli K-12 BL21 (DE-Δtail), called BL21 here for simplicity, which are sensitive to phage T6. The negative control experiments were performed using bacterial cells E.coli K-12 BW25113 with TSX−, called TSX− here, which are insensitive to phage T6. The phages used throughout the experiments were T6 phages.
[0058]Bacterial cultures were prepared overnight using frozen cultures inoculated in luria broth (LB) growth media. The overnight cultures were then inoculated to make fresh samples to be used the same day. The growth of the cells was monitored using a spectrophotometer that read the optical density (OD)...
example 3
[0079]This example demonstrates that lysostaphin can be used to detect the presence of a bacterium in a sample using methods and devices according to the present disclosure. In this example, the lysostaphin from Staphylococcus simulans was combined with Staphylococcus aureus 8325 as the positive control. As negative controls, lysostaphin from Staphylococcus simulans was combined with E.coli K12 BW25113, and lysostaphin from Staphylococcus simulans was combined with Pseudomonas aeruginosa (PA01).
[0080]FIG. 11 shows the results of both positive and negative control experiments. As indicated, the integrated potassium-sensitive chip can detect the presence of the specific bacterial strain Staphylococcus aureus 8325 using the lysostaphin from Staphylococcus simulans. As shown, the output increases as a result of the efflux of potassium ions from cells in the positive control test, as compared to the negative controls.
example 4
[0081]This example demonstrates that antibiotics can be used to detect the presence of a bacterium in a sample using methods and devices according to the present disclosure. In this example, the Polymyxin B antibiotic (PMB) was combined with E.coli K12 BW25113 in SM buffer as the positive control. As the negative control, Polymyxin B antibiotic (PMB) combined with E.coli K12 BL21 (DE3-Δtail) (referred to as BL21) in SM buffer. Experiments were performed as described above for Example 1, using the ISFET system 600.
[0082]FIG. 12 shows the results of both positive and negative control experiments. As indicated, the integrated potassium-sensitive chip can detect the presence of the specific bacterial strain E.coli K12 using the Polymyxin B antibiotic (PMB). As shown, the output increases as a result of the efflux of potassium ions from cells in the positive control test, as compared to the negative control.
[0083]FIGS. 13A-13C illustrate applications for ISFET-based bacteria sensors, in ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com