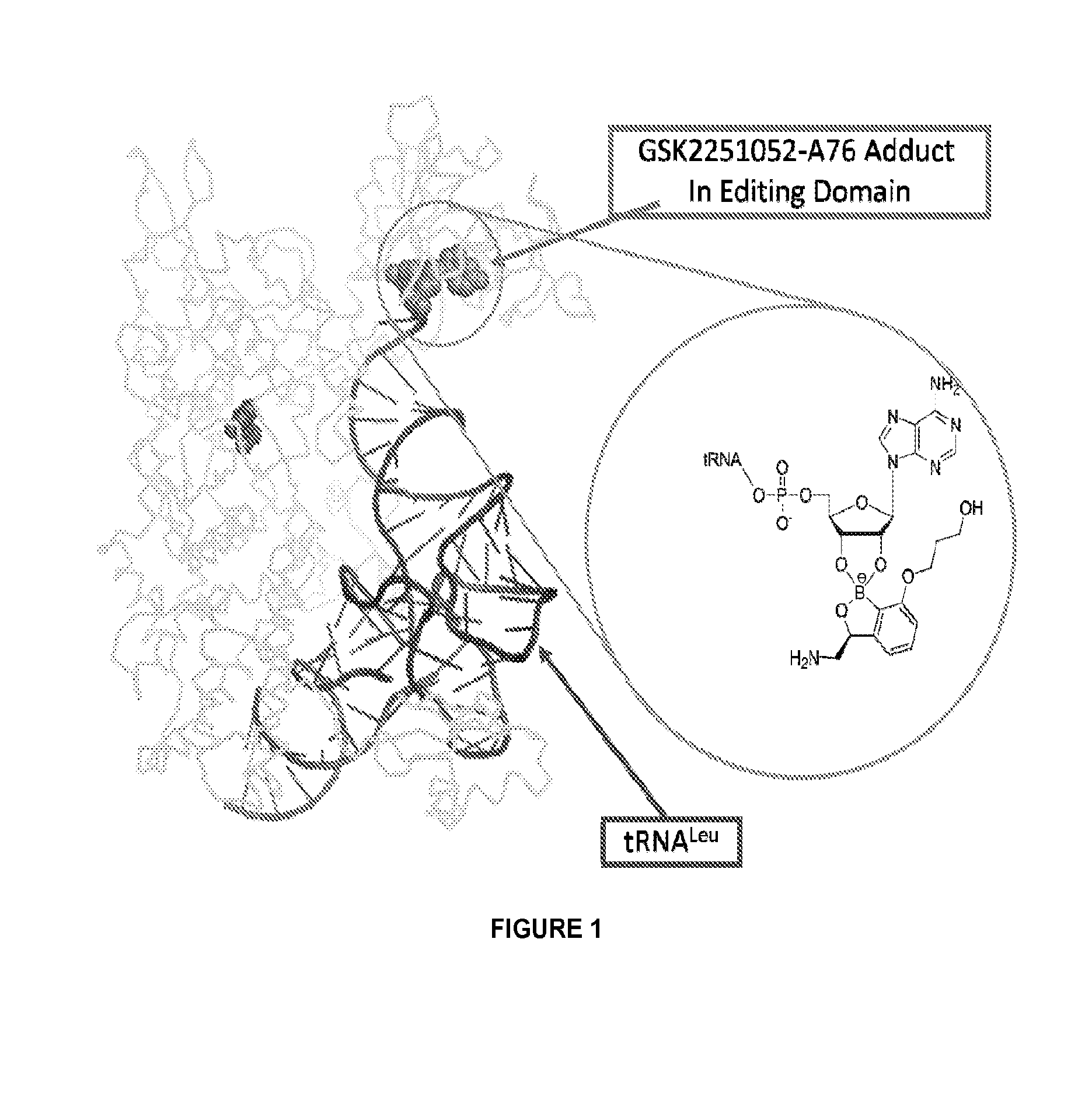
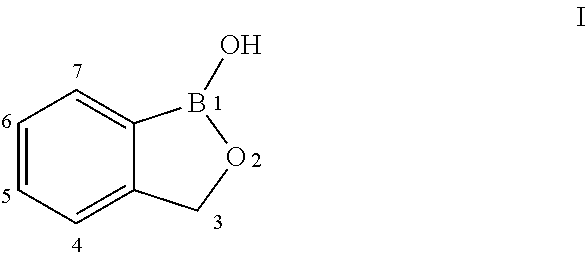
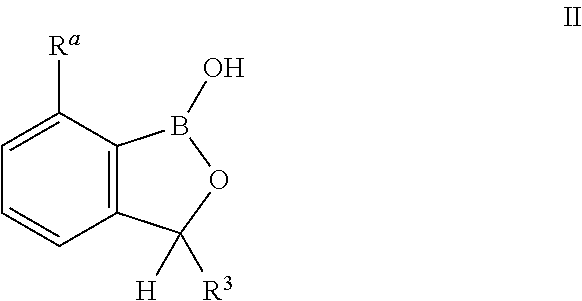
Benzoxaborole compounds and uses thereof
a technology of benzoxaborole and compounds, applied in the field of new boron-containing molecules, can solve the problems of no longer editing mischarged amino acids, affecting the synthesis of proteins, and affecting the synthesis of proteins, and achieve the effect of reducing the resistance ra
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Resistant Mutants to GSK2251052 Generated In Vitro
[0111]Spontaneous Resistance Frequency was determined in vitro for GSK2251052 and the resistance mutants in E. coli ATCC 25922, K. pneumoniae ATCC 13883, and P. aeruginosa ATCC 27853 strains were characterized. GSK2251052 was prepared as described in U.S. Pat. No. 7,816,344 (the entire contents of which are hereby incorporated by reference herein). LB broth, Mueller Hinton II broth (adjusted), and Difco noble agar were obtained from Becton Dickinson (Cockeysville, Md., USA). M9 minimal broth containing 1×M9 salt, 0.5 μg / ML thiamine, 1 mM MgSO4, 0.1 mM CaCl2, 2% glucose, was obtained from Teknova (Hollister, Calif., USA). Antibacterial agents ciprofloxacin, ceftazidime, chloramphenicol, gentamycin, kanamycin, polymixin B, tobramycin and trimethoprim, and amino acids were obtained from Sigma Chemicals (St. Louis, Mo., USA). Strains were obtained from ATCC (Manassas, Va., USA).
[0112]Determination of the Minimum Inhibitory Concentration....
example 2
Resistant Mutants to GSK2251052 Generated In Vivo
[0130]A comparative, dose-ranging study of GSK2251052 vs. imipenem-cilastatin in complicated lower urinary tract infection and pyelonephritis was undertaken with 210 targeted patients (20 actually enrolled) in a phase II clinical trial. The study was designed to determine the appropriate and safe dose for Phase III trials. Doses were 750 mg or 1500 mg twice a day, compared to an active comparator, with an independent safety review committee.
[0131]The phase II clinical trial identified four subjects who developed spontaneous resistance to GSK2251052 as evidenced by increased MICs (>32-fold) determined for the clinical isolates from the four subjects. Two of the resistant mutant strains identified were E. coli strains, one was a P. mirabilis strain, and one was a K. pneumoniae strain. The rates of spontaneous resistance for the baseline isolates tested were not significantly different from standard strains previously tested (˜10−7).
[013...
example 3
Measurement of the Suppression of the Emergence of a Resistant Bacterial Strain In Vitro
[0142]Clinical strains of Escherichia coli and Proteus mirabilis isolated from the above-described clinical trial study (Table 12), along with other E. coli strains were used to measure the suppression of the emergence of resistant bacterial strains in vitro and to determine the susceptibility of these GSK2251052 resistant bacterial isolates to norvaline. Increased sensitivity to this leucine analog would suggest an editing domain mutation in LeuRS. Moreover, in order to find out if sub-inhibitory concentrations of norvaline could suppress growth of GSK2251052 resistant mutants under restricted amino acid conditions in the laboratory, the frequency of spontaneous resistance to GSK2251052 was determined for Escherichia coli 1162222 (standard strain) and the E. coli isolate (baseline strain from subject E. coli 1, isolated from blood in the above-described clinical study in Example 2) in the presen...
PUM
| Property | Measurement | Unit |
|---|---|---|
| volume | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
| concentration | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com