Method for detecting cells
a cell identification and cell technology, applied in the field of cell identification, can solve the problems of laborious and inefficient, inherent experimental variability of the current method used to detect/isolate pluripotent cells, and low efficiency of the current method, so as to achieve the effect of opening new possibilities for identification
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
Nucleosomes in Interphase Nuclei of Human Somatic Cells are Organized in Discrete Nanodomains
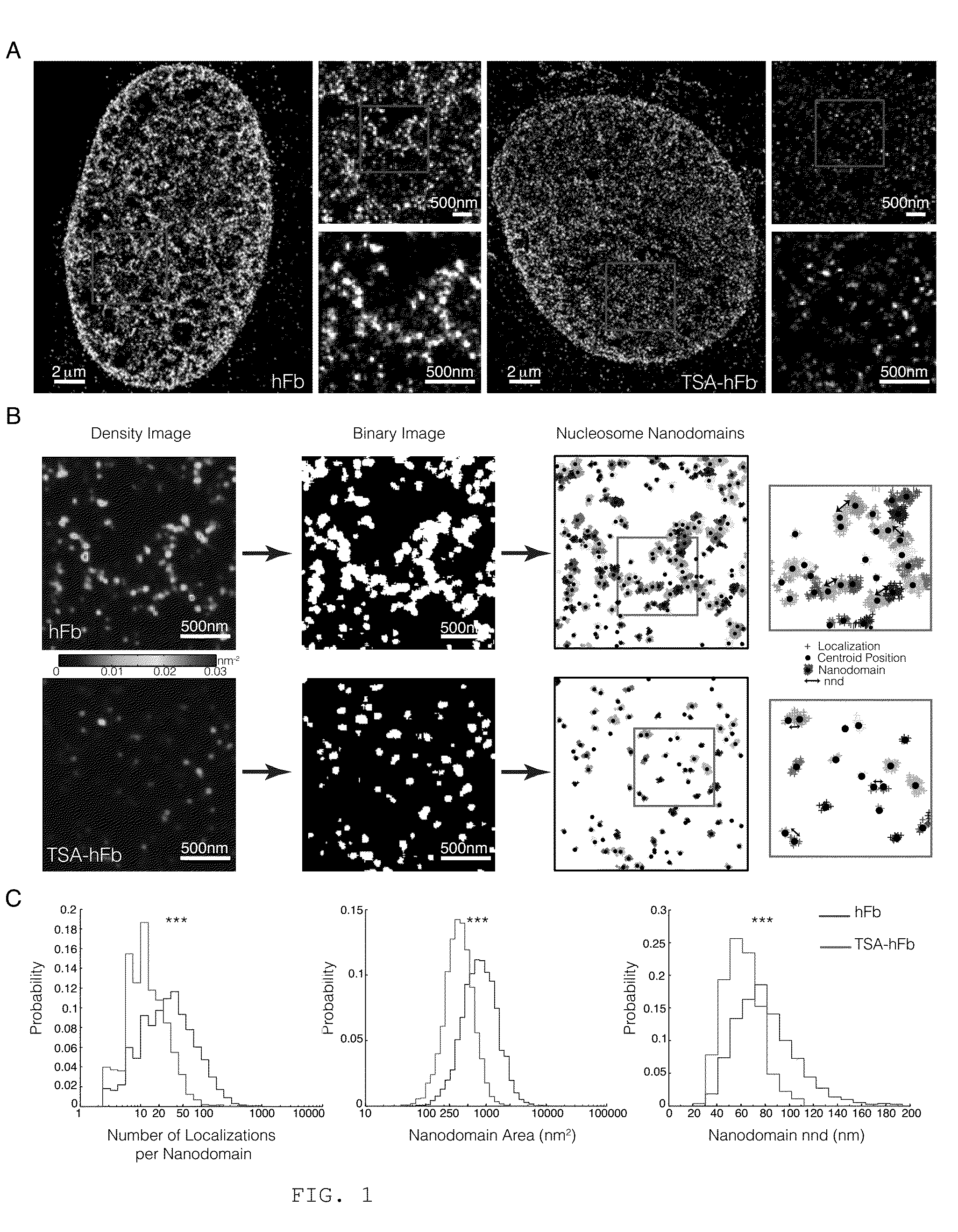
[0215]To reveal the organization of chromatin at nanoscale resolution, the inventors recorded STORM images of the core histone protein H2B in interphase human fibroblast nuclei (hFb). An antibody that recognizes native H2B was used. STORM images revealed a striking organization of H2B inside the nucleus (FIG. 1A, left), which was not evident with conventional fluorescence microscopy (FIG. 7A). H2B appeared clustered in discrete and spatially separated nanodomains (FIG. 1A, right zooms). The H2B nanodomain density (number of nanodomains per unit area) was ˜25% higher in the nuclear periphery, where the heterochromatin is thought to be located, compared to the nuclear interior. Since H2B is a core histone of the nucleosome octamer, its localization should reflect the arrangement of nucleosomes within the chromatin fiber. In accordance with this idea, another core histone protein of the nucleos...
example 2
Nucleosomes in Interphase Nuclei of Mouse Embryonic Stem Cells Form Discrete Nanodomains, Whose Organization Correlates with Ground State Pluripotency
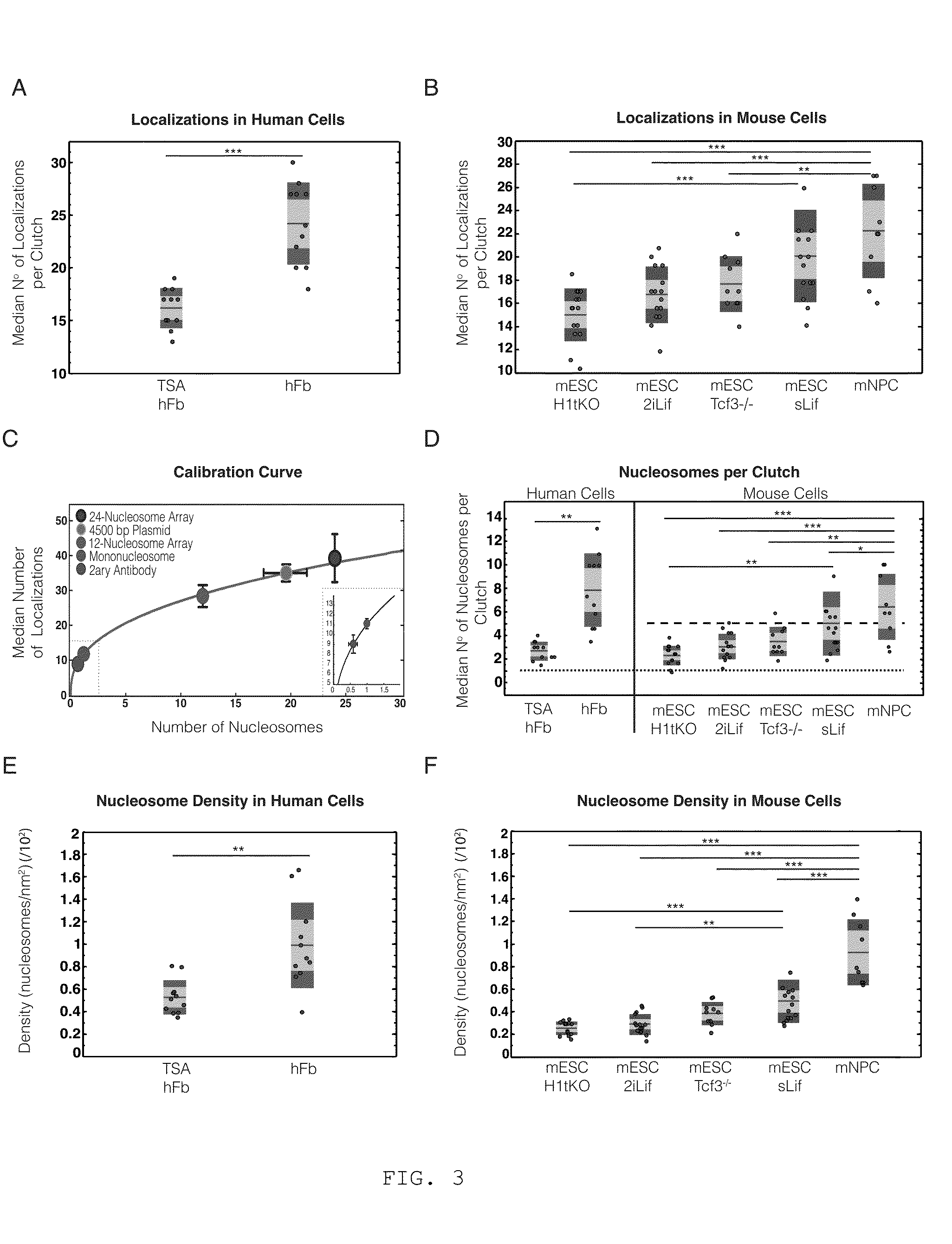
[0219]To assess the H2B organization of pluripotent stem cells, the inventors next imaged mouse embryonic stem cells (mESC) with STORM. mESCs were initially cultured in a medium containing serum and the Leukemia inhibitory factor (sLif) and H2B was labeled by immunofluorescence and imaged with STORM as before. STORM images of these mESCs showed two different categories of nuclei. The first category, Type 1, displayed nanodomains that appeared bright (i.e. contained a large number of localizations) similar to hFbs (FIG. 2A, yellow arrowheads). The second category, Type 2, on the other hand, displayed an increased amount of dimmer nanodomains (i.e. containing a small number of localizations) and the nanodomains were more dispersed within the nucleus, similar to TSA-hFbs (FIG. 2B, cyan arrowheads). It has been reported that mESCs culture...
example 3
Nanodomains Contain a Discrete Number of Nucleosomes and the Nucleosome Number Correlates with Cell Pluripotency
[0223]The inventors next aimed to further quantify the changes they observed in the number of localizations (and hence brightness) of nanodomains in terms of the number of nucleosomes. There is not a one-to-one relationship between the number of localizations in STORM images and the number of nucleosomes mainly for two reasons: i) the antibody epitope labeling efficiency may not be 100%, ii) each fluorophore can undergo multiple photoswitching events, resulting in multiple localizations arising from a single fluorophore. However, the epitope labeling efficiency of the H2B antibody should be comparable across the human cells (hFbs, TSA-hFbs) and likewise across the different mESCs analyzed. In addition, the antibodies used were always labeled with a similar dye composition (Extended Experimental Procedures) and each cell was imaged in the same way (Table I) to obtain compar...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com