Methods for determining the efficacy profile of a drug candidate
a drug candidate and efficacy profile technology, applied in the field of methods for determining the efficacy profile of a drug candidate, can solve the problems of inability to translatability the efficacy and toxicity profiles of drug candidates from rodent species to humans, and the use of nhp species in drug discovery remains a controversial matter
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
eotide Activity in Mouse Primary Neuronal Cell Cultures
[0310]Oligonucleotides targeting the part of SNHG14 long non-coding RNA which is antisense to the UBE3A pre-mRNA (position 55319 to 141053 of SEQ ID NO: 1) were tested for their ability to reduce the SNHG14 long non-coding RNA transcript preventing UBE3A expression (also termed UBE3A suppressor or UBE3A-SUP in the data table) and their ability to induce UBE3A mRNA re-expression in mouse primary cortical neuron cell cultures, obtained as described in the “Materials and methods” section above. The oligonucleotide concentration was 5 microM. The oligonucleotides were screened according to the protocol for screening in mouse cortical neuron cell cultures described in the section “Materials and methods”. The results are shown in table 4.
TABLE 4Oligonucleotide activity in primary mouse neuronal cell cultures.CMP% of Mock% of MockID NOoligonucleotideUBE3A_SUPsdUBE3Asd 95_1CTCAtacttgctttaAT 3.6 0.1154.115.1 95_2CTcatacttgctttaAT15.9 2.6...
example 2
eotide Activity in Human Neuronal Cell Cultures
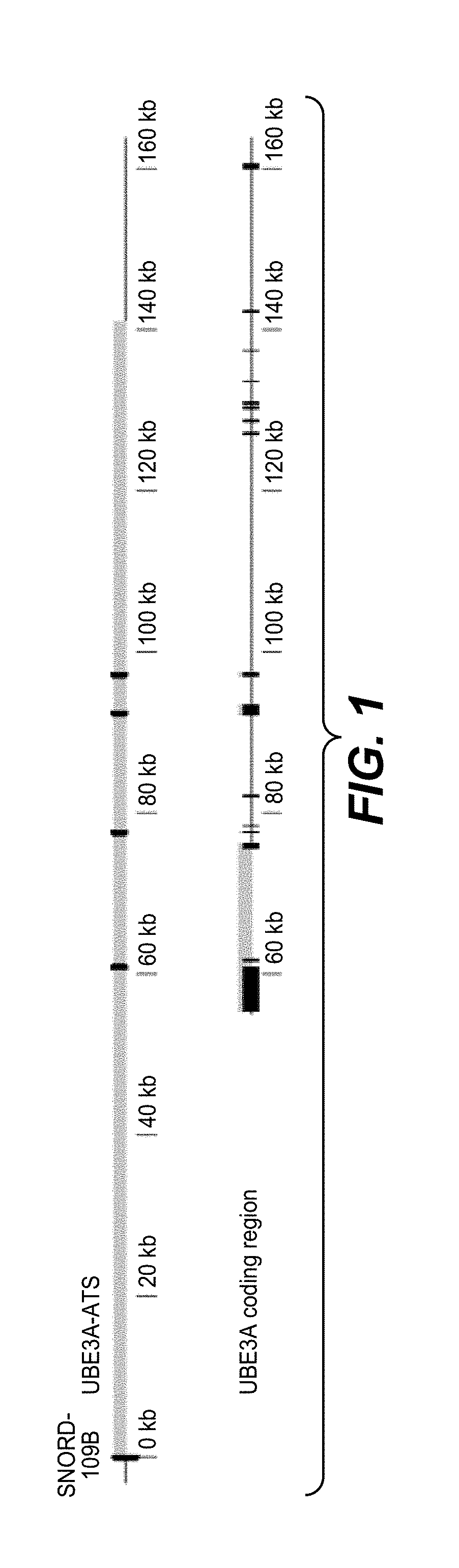
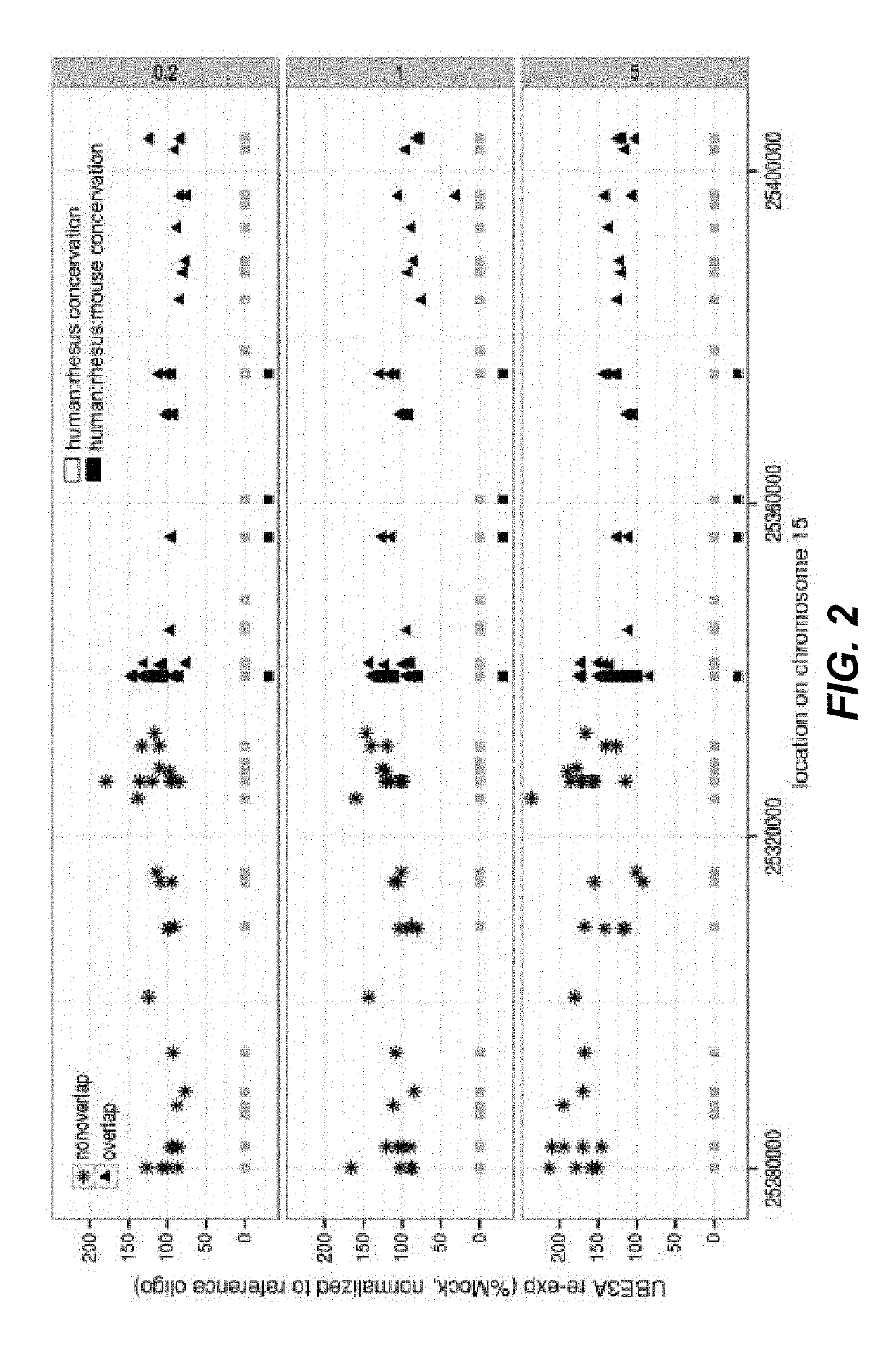
[0311]Oligonucleotides targeting human SNHG14 in the region downstream of SNORD109B corresponding to position 25278410 to 25419462 on chromosome 15 (SEQ ID NO: 1) were tested in patient derived human neuronal cell cultures (see protocol in “Materials and methods” section). The oligonucleotides ability to reduce the SNHG14 transcript in the region downstream of SNORD109B (also termed UBE3A suppressor or UBE3A-SUP in the data table), without affecting expression of SNORD115 was analyzed. Furthermore, the ability to induce UBE3A mRNA re-expression was analyzed.
[0312]The oligonucleotides were screened according to the protocol for screening oligonucleotides in human neuronal cell cultures described in the section “Materials and methods” above.
[0313]The results are shown in table 5. The expression of UBE3A mRNA has been measured for all compounds, whereas the knock-down of the UBE3A suppressor and the maintenance of SNORD1115 levels have not...
example 3
n and Validation of Cyno Neuronal Differentiation In Vitro System
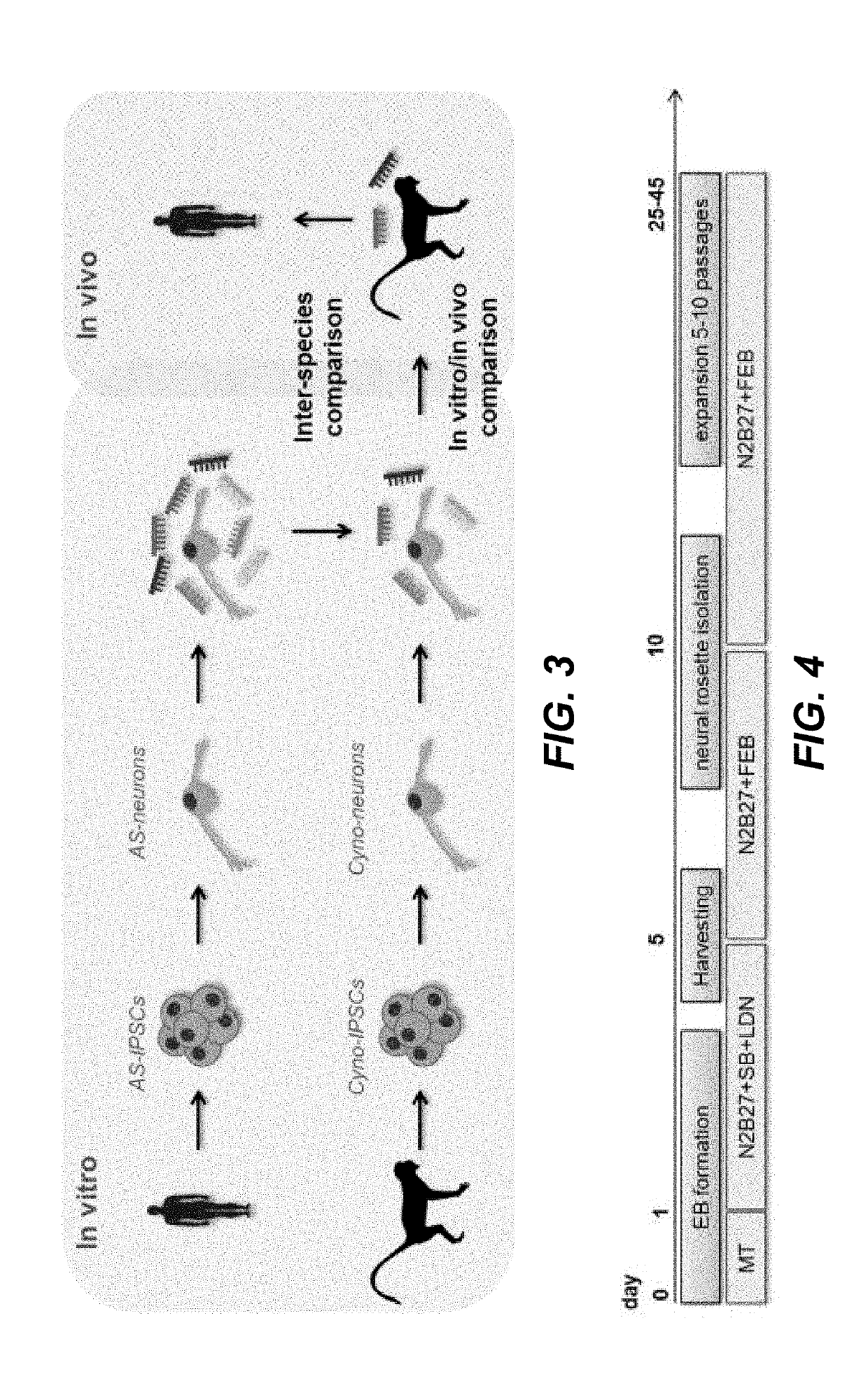
[0316]Cyno IPSCs were induced to differentiate into NPCs following the protocol as described in the Materials and Methods section and depicted in FIG. 4. The protocol allows to derive NPCs which can be maintained and expanded in basal medium supplemented with FGF, EGF and BDNF. To verify the efficient induction of cyno NPCs, expression of the neural stem cell markers SOX2 and NESTIN was evaluated by immunostaining (see FIG. 5A and FIG. 5B). Cyno NPCs express SOX2 and NESTIN and the expression pattern is highly comparable to human NPCs. To derive differentiated NCs, expanding NPCs are dissociated and plated in SFA medium for a week. Afterwards the cells are exposed to differentiation medium (BGAA). To evaluate the neuronal differentiation potential of cyno NPCs, a transcriptional analysis was performed and results were compared to the transcriptional profile of differentiated human NPCs (see FIG. 6A, FIG. 6B, FIG. 6C, F...
PUM
| Property | Measurement | Unit |
|---|---|---|
| nucleic acid sequence | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com