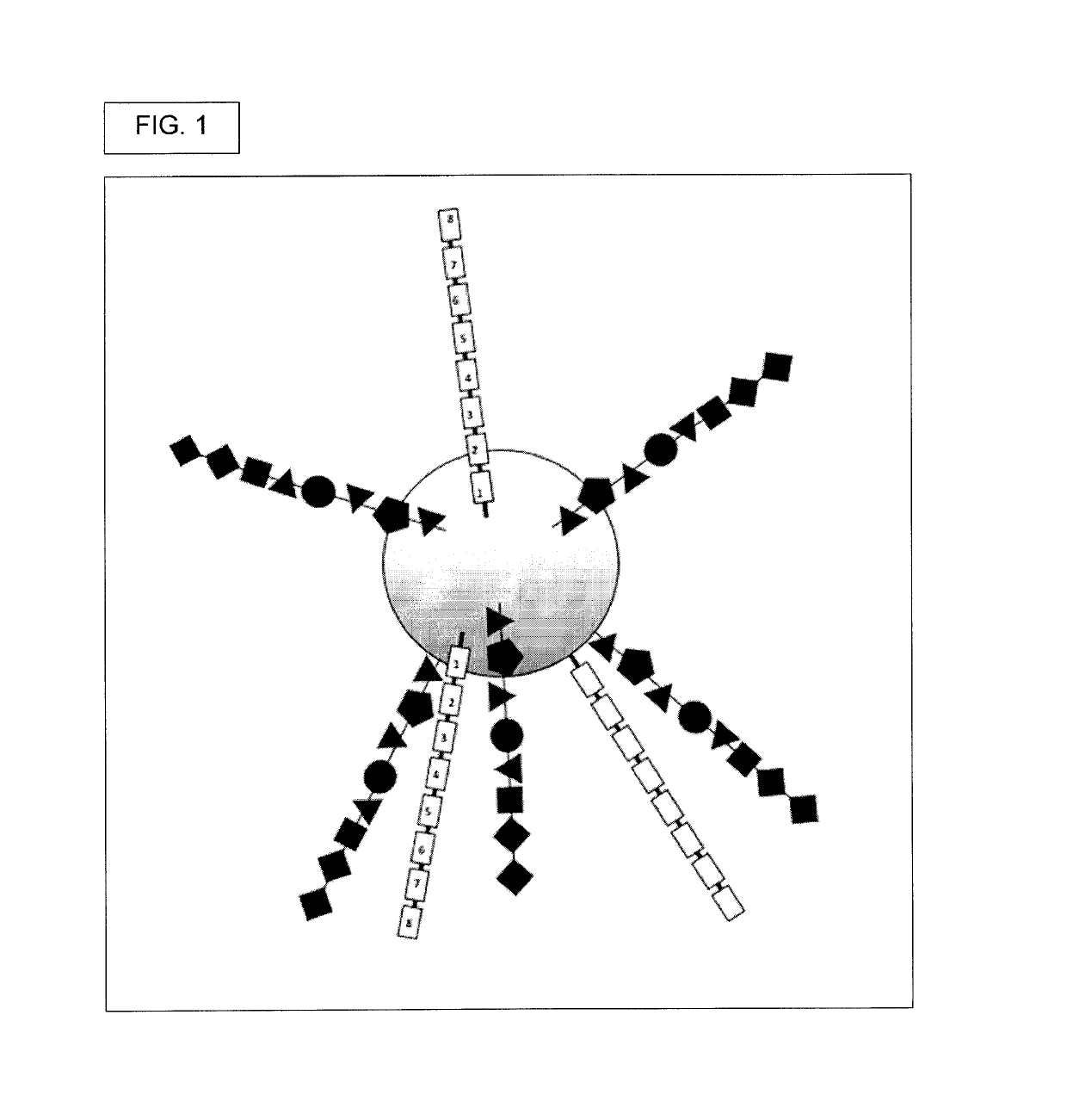
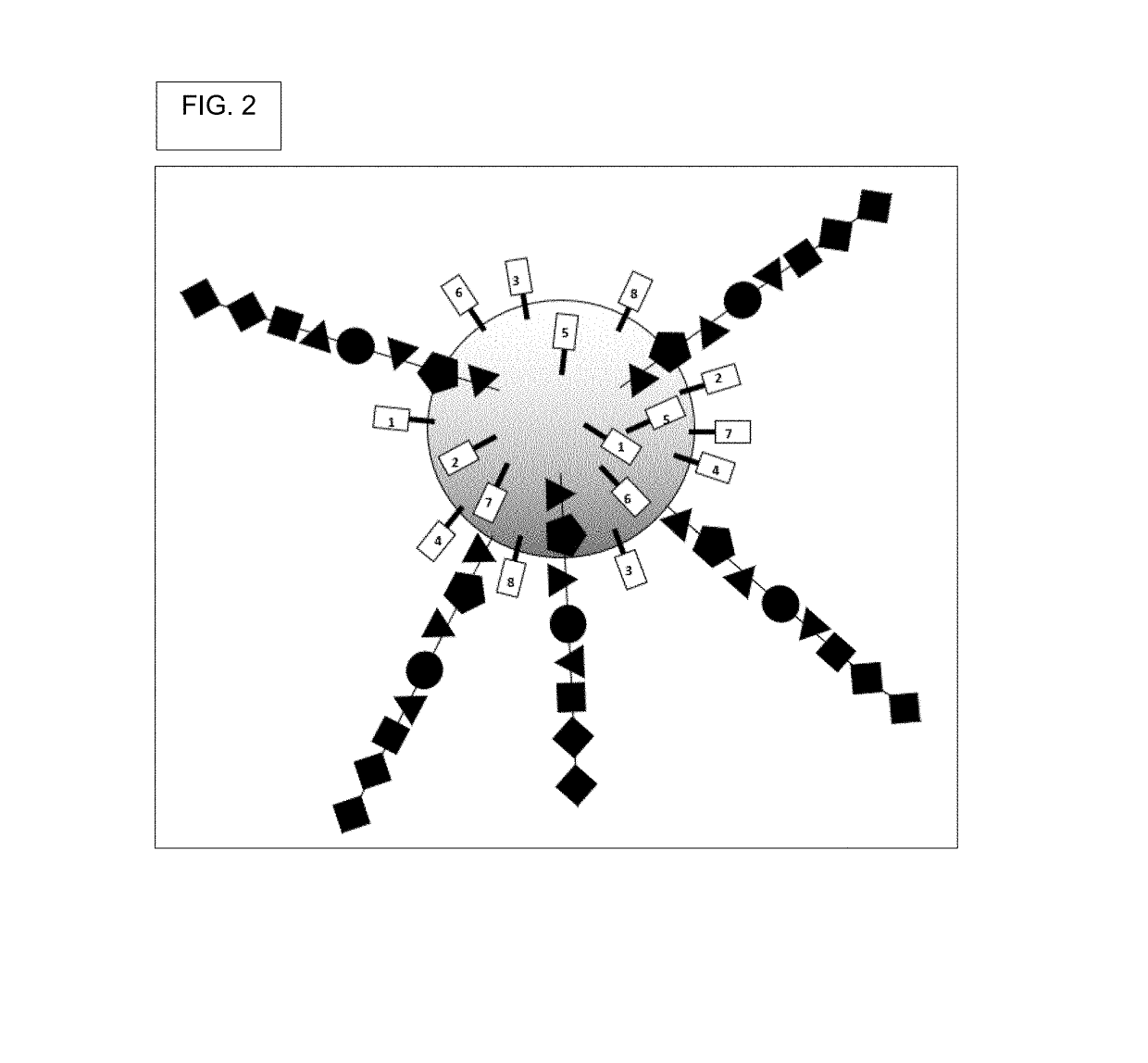
Oligonucleotide encoded chemical libraries
a technology of oligonucleotide and chemical library, applied in library screening, library creation, laboratory glassware, etc., can solve the problems of high cost, inability to pre-select a target, inefficient drug screening method, etc., and achieve the effect of minimizing or preventing fluid leakage, and minimizing or preventing fluid evaporation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Benefits of technology
Problems solved by technology
Method used
Image
Examples
example 1
kflow
[0498]The present disclosure provides methods, including that outlined below as “First Workflow” and as “Second Workflow.”
The First Workflow includes the steps: (1) Generate DELB, (2) Beads into picowells, (3) Load assay reagents into picowells, (4) Release bead-bound compounds, (5) Measure assay readout, (6) Rank the assay readout, and (7) Generate a new set of DELBs.
[0499]Generate DELB. First, create the DNA encoded library on beads (DELB). Each bead contains a population of the exact, same compound, though slight departures from this may occur where some of the manufactured compounds had incomplete couplings or were suffered chemical damage, such as inadvertent oxidation.
[0500]Beads into picowells. Then, deposit beads in picowells. In a preferred embodiment, each picowell gets only one bead. Each picowell can have a round upper edge, a round lower edge, a solid circular bottom, an open top, and a wall. The wall's bottom is defined by the round upper edge and by the round low...
example two
flow
[0509]The Second Workflow involves picowells that are sealed with caps. The caps can take the form of spheres of slightly greater diameter than the diameter of the picowells, where this diameter is measured at the top rim of the picowell (not measured at the bottom of the picowell). The cap can be made to fit snuggly into the top of the picowell by subjecting the entire picowell plate to mild-gravity centrifugation. In Second Workflow, the caps take the form of beads that contain linkers, where each linker is linked to a compound. The linkers are cleavable linkers, where cleavage released the compounds and allows them to diffuse to the cells. This type of cap is called an “active cap.” The Second Workflow includes the steps, (1) Generate DELB, (2) Load assay reagents into picowells, (3) Cap picowells with DELB, (4) Release bead-bound compounds from the bead that acts as a cap, (5) Measure assay readout, (6) Determine sequence of the DNA barcode that is on the bead; (7) Rank the ...
example 3
ontrol
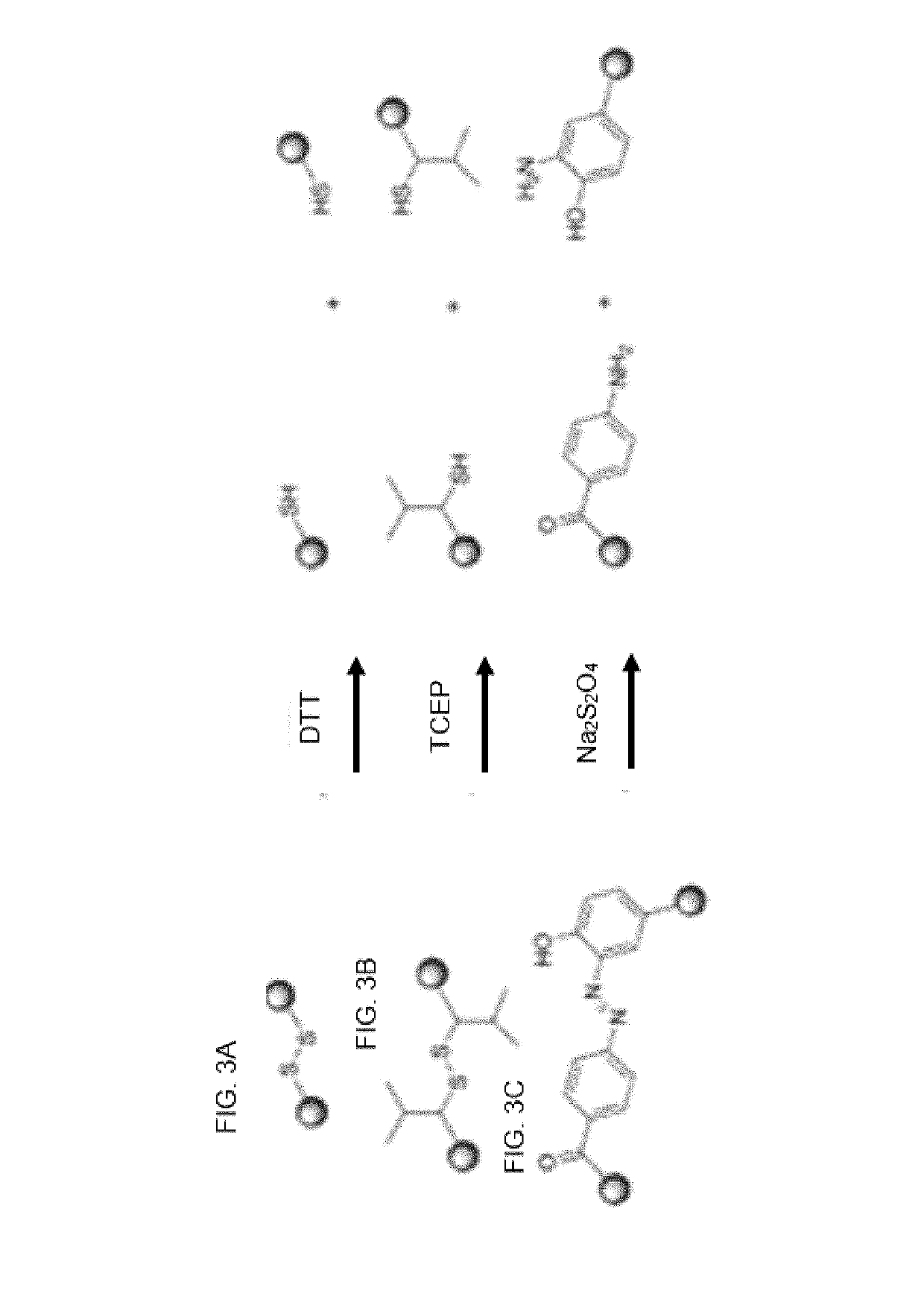
[0510]This concerns controlling and monitoring release of bead-bound compounds. Applicants devised the following procedure for synthesizing bead-bound release-monitor. See, FIG. 11 and the following text.
[0511]FIG. 11 describes steps in the organic synthesis of the above exemplary embodiment of a bead-bound release-monitor.
[0512]Step 1. Provide the Resin
[0513]TentaGel® resin (M30102, 10 μm NH2, 0.23 mmol / g, 10 mg; MB160230, 160 μm RAM, 0.46 mmol / g, 2 mg) was weighed into a tube (1.5 mL Eppendorf) and swelled (400 μL, DMA).
[0514]Resin was transferred into fritted spin-column (MoBiCol® spin column, Fisher Scientific), solvent removed through filter by vacuum, and pendent Fmoc was deprotected (5% Piperazine with 2% DBU in DMA, 400 μL; 2×10 min at 40° C.). The MoBiCol spin column has a 10 micrometer large frit and a luer-lock cap.
[0515]Resin was filtered over vacuum, and washed (2×DMA, 400 μL; 3×DCM, 400 μL; 1×DMA, 400 μL).
[0516]Step 2. Couple Lysine Linker to Resin
[0517]A solutio...
PUM
| Property | Measurement | Unit |
|---|---|---|
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
| size | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com