Rna-dependent rna polymerase, methods and kits for the amplification and/or labelling of rna
An RNA polymerase, kit technology, applied in biochemical equipment and methods, enzymes, transferases, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0211] Preparation method of recombinant norovirus-RdRP.
[0212] The cDNA of Norwalk virus-RdRP was obtained by performing PCR on the clone pUS-NorII (GenBank accession number: AY741811) of Norwalk virus. The cDNA was cloned into the pET-28b(+) vector (Novagen), and the expression vector was sequenced and transformed into E. coliCL21(DE3)pLysS. Cells were incubated at 37°C in Luria-Bertani medium containing kanamycin (50 mg / l). Protein expression was induced by adding isopropyl-β-D-thiogalactoside (IPTG) to a final concentration of 1 mM at an optical density of 0.6 (OD600). Then incubate overnight at a temperature of 25°C. Cell extracts obtained from 250 ml of culture were washed once with 4 ml of phosphate buffered saline (PBS) and 1% Triton X 100 (sigma) water. Cells were treated with DNase enzyme (10 U / ml) at 37° C. for 15 minutes, sonicated, and suspended in 40 ml of binding buffer (20 mM Tris / HCl, pH 7.9, 500 mM NaCl, 5 mM imidazole). Centrifuge at 4300 rpm for 40 mi...
Embodiment 2
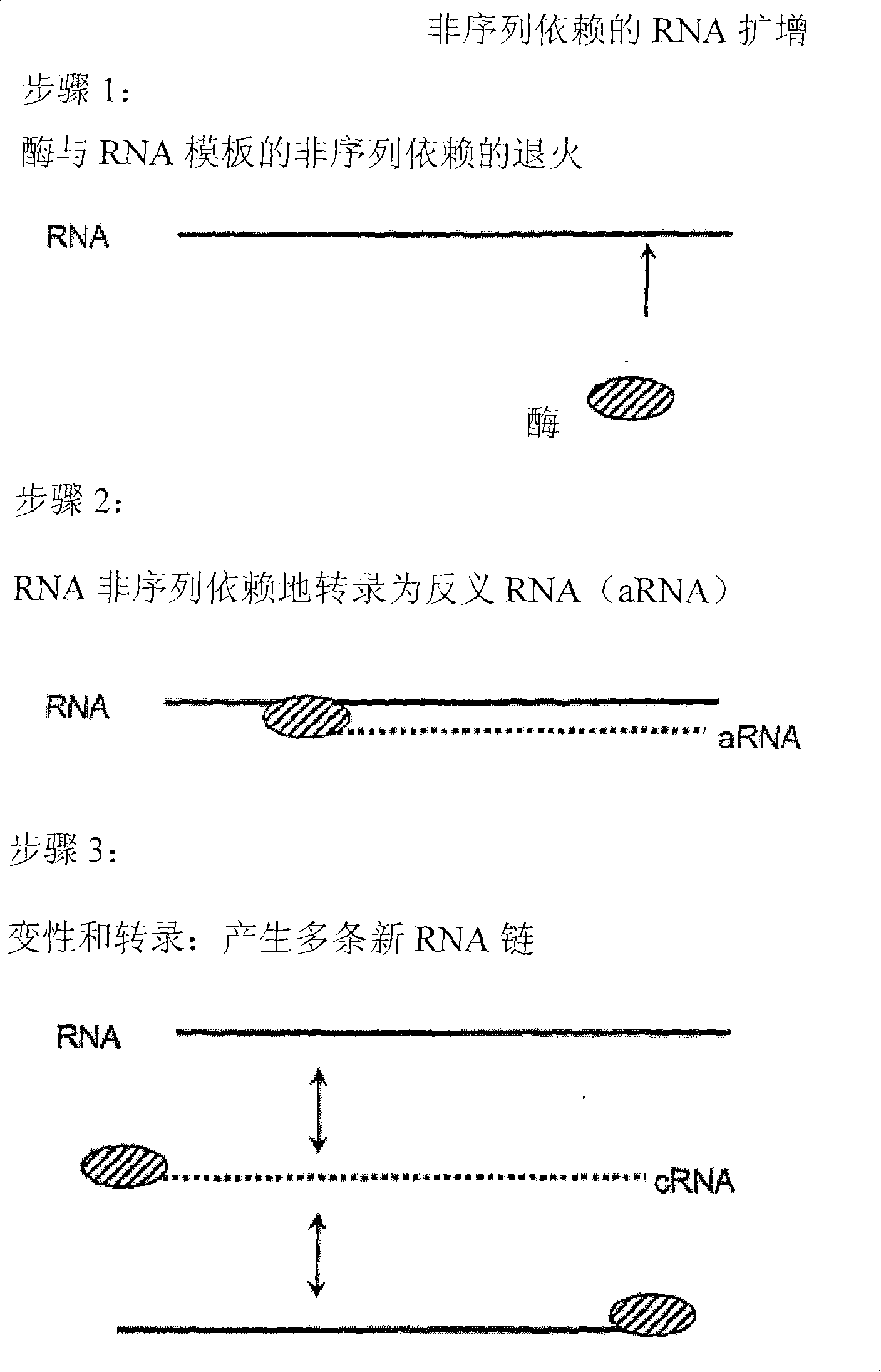
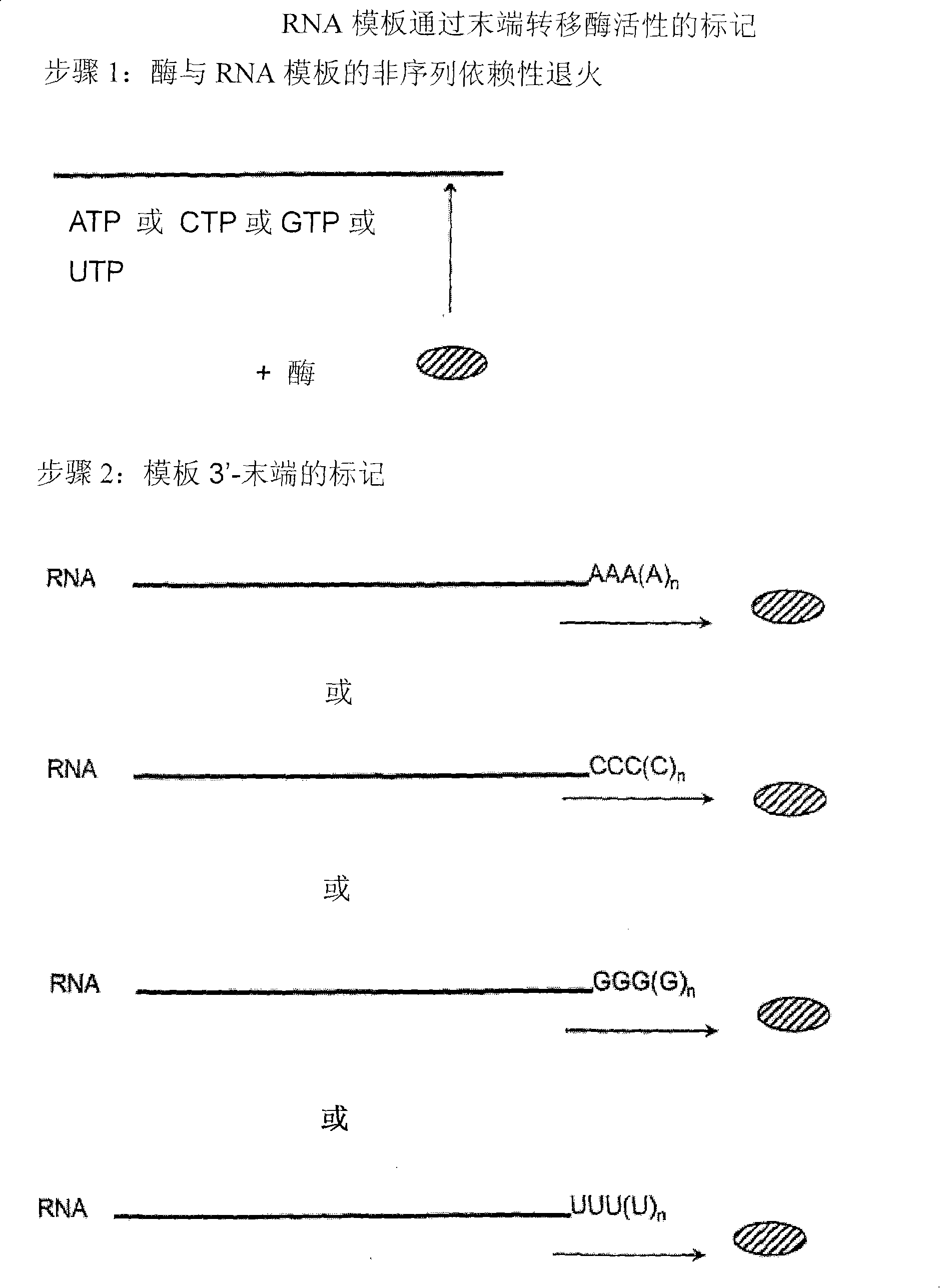
[0214]Sequence-independent RNA amplification using Norwalk virus-RdRP.
[0215] 0.5-1 μg RNA template, 10 μl reaction buffer (250mM HEPES, pH 8.0, 15mM magnesium acetate, 20mM DTT), 50U RNase inhibitor (RNAsin, Promega), ATP, CTP, GTP, UTP each 0.4mM, Example 1 A reaction mixture (50 μl) consisting of 3 μM norovirus-RdRP prepared in , was reacted at 30° C. for 2 hours. The reaction was terminated by adding 50 μl of stop solution (4M ammonium acetate, 100 mM EDTA). Purification was carried out by phenol-chloroform extraction, or by MEGAclear kit (Ambion) according to the manufacturer's instructions. After staining with ethidium bromide, the transcripts were observed on agarose gel prepared in TBE buffer by UV transmission illumination. Formamide agarose gels can also be used.
Embodiment 3
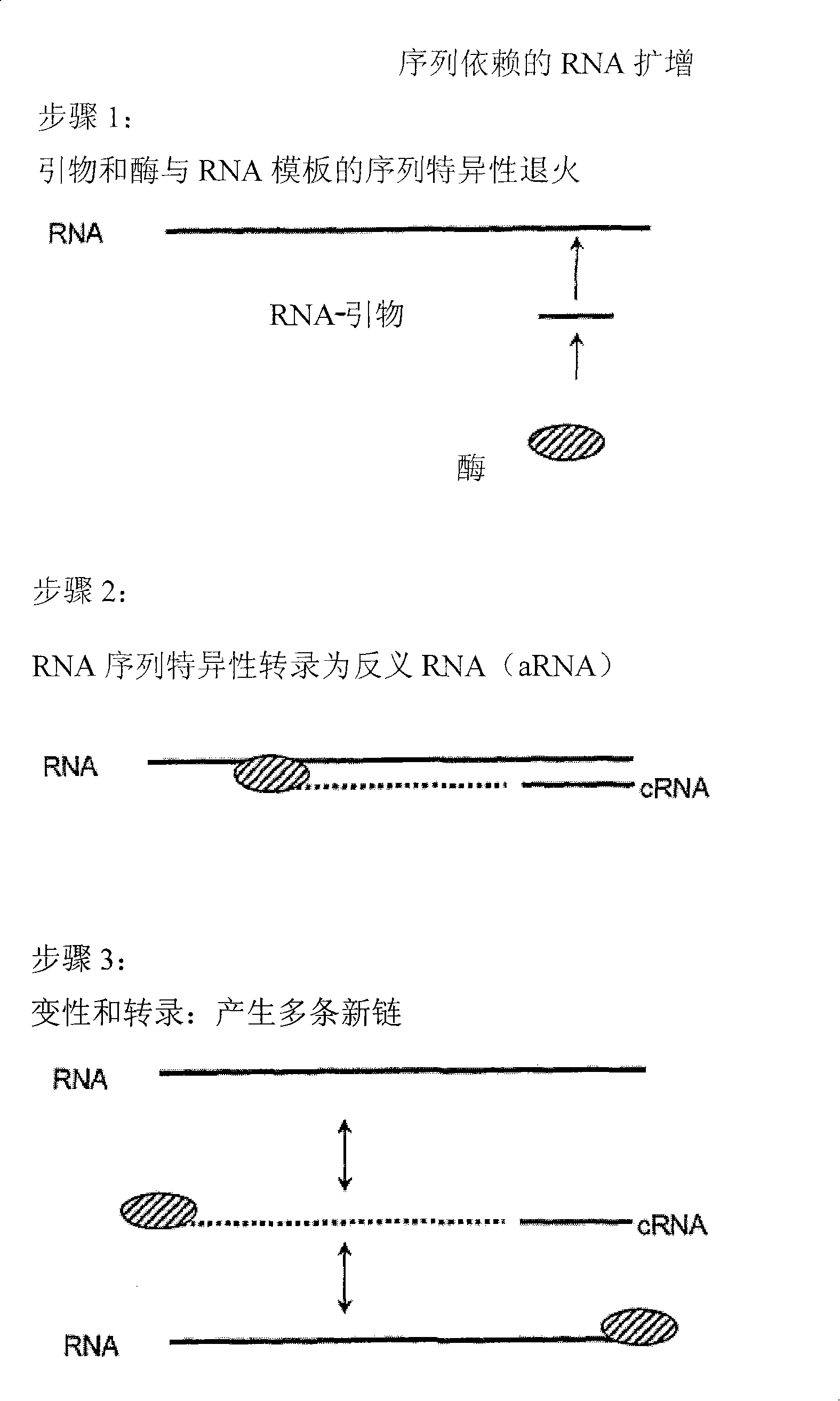
[0217] Sequence-dependent RNA amplification using the gene-specific RNA primer, Norwalk virus-RdRP.
[0218] 0.5~1μg RNA template, 10μl reaction buffer (250mM HEPES, pH 8.0, 15mM magnesium acetate, 20mM DTT), 50U RNase inhibitor (RNAsin, Promega), ATP, CTP, GTP, UTP each 0.4mM, 0.1~1μM A reaction mixture (50 μl) consisting of gene-specific RNA primers and 3 μM Norwalk virus-RdRP prepared in Example 1 was reacted at 30° C. for 2 hours. The reaction was terminated by adding 50 μl of stop solution (4M ammonium acetate, 100 mM EDTA). Purification was carried out by phenol-chloroform extraction, or by MEGAclear kit (Ambion) according to the manufacturer's instructions. After staining with ethidium bromide, the transcripts were observed on agarose gel prepared in TBE buffer by UV transmission illumination. Formamide agarose gels or urea / polyacrylamide gels can also be used.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com