Mass spectrum reagent kit and method for detecting and prognosis judging CEA negative gastric cancer
A detection kit and the technology of the kit, which are applied in the field of protein detection, can solve the problems of lack of early monitoring methods in the prognosis evaluation of CEA-negative gastric cancer patients, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0078] Example 1 Distinguishing between normal and CEA-negative gastric cancer patients and preparation of a mass spectrometry kit
[0079] (1) Experimental method
[0080] The preoperative sera of 280 patients with CEA-negative gastric cancer were collected, with an average age of 47 years. 280 healthy subjects, with an average age of 45 years old, were from a population with normal liver and kidney function tests. Collect 1mL of venous blood from the subject on an empty stomach, immediately after collection, let it stand in a refrigerator at 4°C for 2 hours, centrifuge at 4,000r / min at 4°C for 10 minutes to separate the serum, and centrifuge the serum again at 12,000r / min at 4°C for 5 minutes to remove all residual cell debris and insoluble matter, the serum was divided into 100 μL / tubes on ice, a total of 5 tubes, and stored in a -80°C refrigerator. Avoid repeated freeze-thaw cycles.
[0081] Protein chip and magnetic beads operation steps
[0082] Serum sample processi...
Embodiment 2
[0095] The double-blind test of embodiment 2 kit
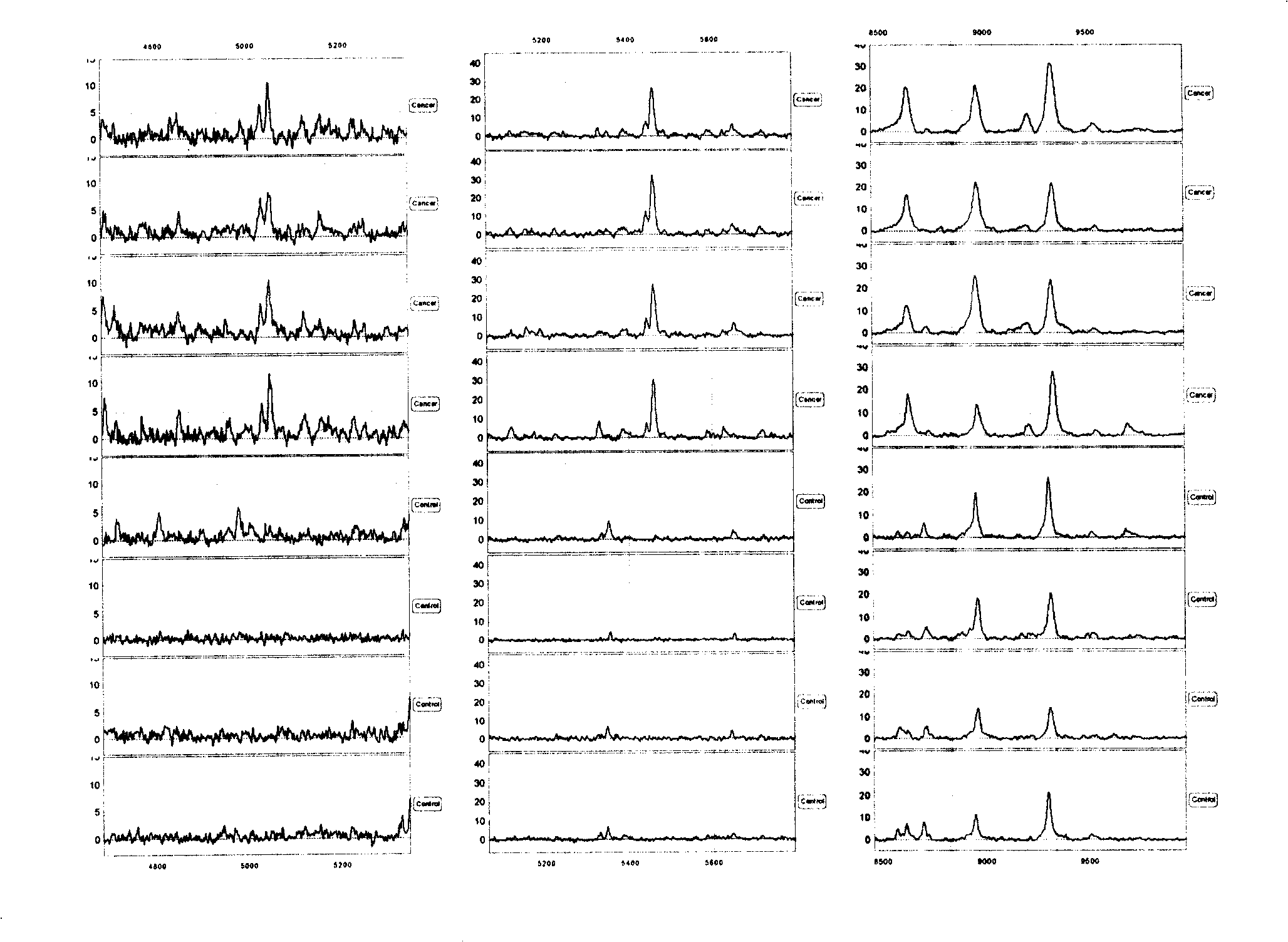
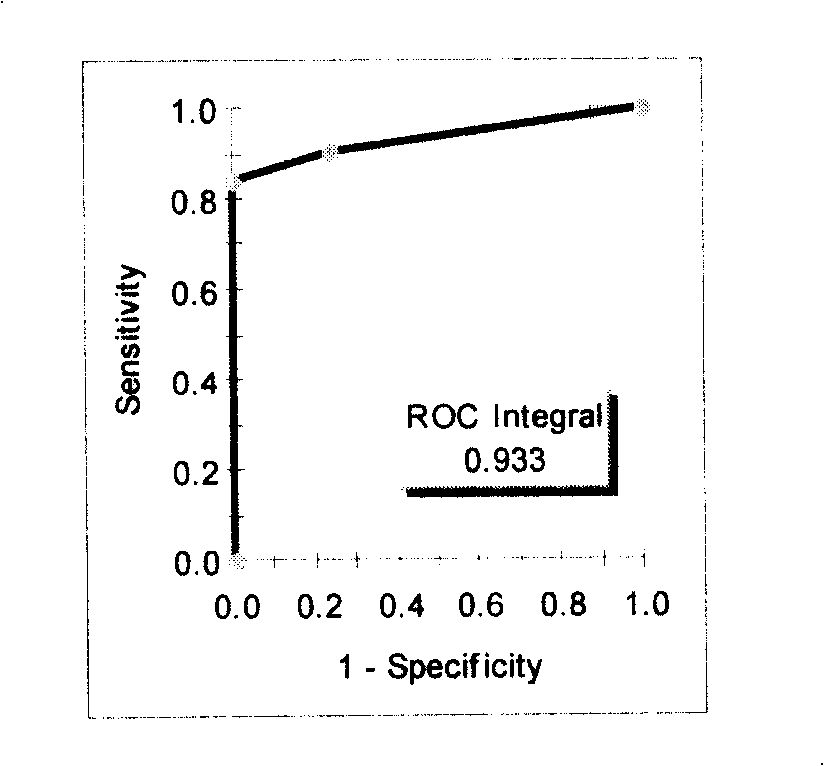
[0096] Screen out several characteristic protein peaks from embodiment 1, the test model that 5047,5461,8628 ± 15Da 3 difference peaks form ( figure 1 The top four cases are gastric cancer patients, and the bottom four cases are healthy people) Blind screening test and ROC curve were used to analyze the mass spectrometry results of 71 CEA negative gastric cancer samples with CEA negative gastric cancer patients and healthy people, and 64 of them were correctly distinguished , 7 were misclassified, and the sensitivity was 90%; 69 of 71 control samples were correctly distinguished, 2 were misclassified, and the specificity was 97% (see Table 2, figure 2 ):
[0097] Table 2: Discrimination of test models in biological samples
[0098]
[0099] Note: Sensitivity 90% (64 / 71); Specificity 97% (69 / 71)
[0100] The experimental results using C8 and C18 hydrophobic matrix magnetic beads or chips are consistent with the experime...
Embodiment 3
[0101] Example 3 Sequencing and identification of 1465.6Da protein
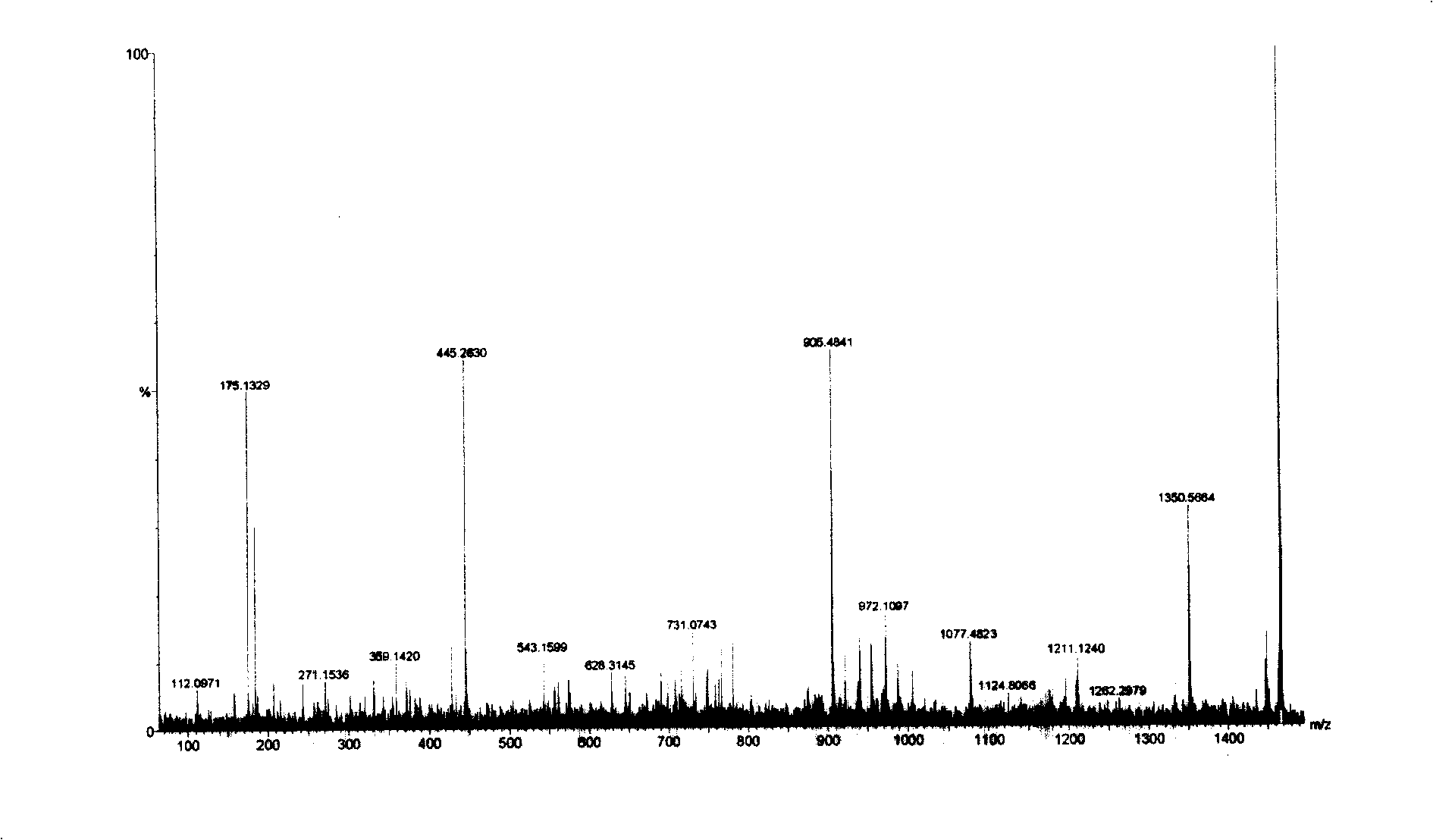
[0102] The 1465.6Da biomarkers were sequenced using multiple-stage mass spectrometry (MS / MS), post-source fragmentation (PSD) and protein ladder sequencing. By breaking molecules into pieces, protein ladders can be generated. This gradient is then analyzed by mass spectrometry. The 1465±1Da protein was identified as a variant fibrinopeptide A (Fibrinopeptide A with alanine truncation at N-terminal) ( image 3 ). Its chemical structure is (15 amino acids arranged from N-terminal to C-terminal):
[0103] N-terminal Asp-Ser-Gly-Glu-Gly-Asp-Phe-Leu-Ala-Glu-Gly-Gly-Gly-Gly-Val-Arg C-terminal.
[0104] Check the chemical structure of the normal fibrinopeptide A molecule in the known genome or cDNA library database (16 amino acids arranged from the N-terminal to the C-terminal, the molecular weight is 1536Da):
[0105] N-terminal Ala-Asp-Ser-Gly-Glu-Gly-Asp-Phe-Leu-Ala-Glu-Gly-Gly-Gly-Gly-Val-Arg C-terminal.
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| Sensitivity | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com