Method for preparing N-terminated acetylated thymosin alpha 1 and special engineering bacteria therefor
A technology of thymosin and engineering bacteria, which is applied in the field of preparing N-terminal acetylated thymosin α1, can solve the problem that a single subunit has no catalytic acetyl transfer activity, and achieve the effect of broad application prospects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0072] Example 1. Expression and acetylation modification identification of fusion protein between thymosin α1 (Tα1) and L12, S18
[0073] 1. Expression of fusion protein of Tα1 and L12 and identification of acetylation modification
[0074] 1. Construction of engineering strain I expressing fusion protein of Tα1 and L12
[0075] The acquisition of the fusion gene: using the Escherichia coli genome as a template, using L1 and L2 as primers to PCR amplify the sequence of the gene rplL encoding ribosomal protein L12, and the product is about 370bp; using the above-mentioned 370bp PCR product as a template, using Ta2 and L2 as primers Primers were used for PCR amplification to obtain a product of about 320bp; then the product of 320bp was used as a template, and Ta1 and L2 were used as primers for PCR amplification to obtain a product of about 370bp, which was the fusion coding gene of the fusion protein of Tα1 and L12, and sequenced , and its nucleotide sequence is shown in seq...
Embodiment 2
[0113] Example 2. Screening of genes related to acetylation modification of Tα1
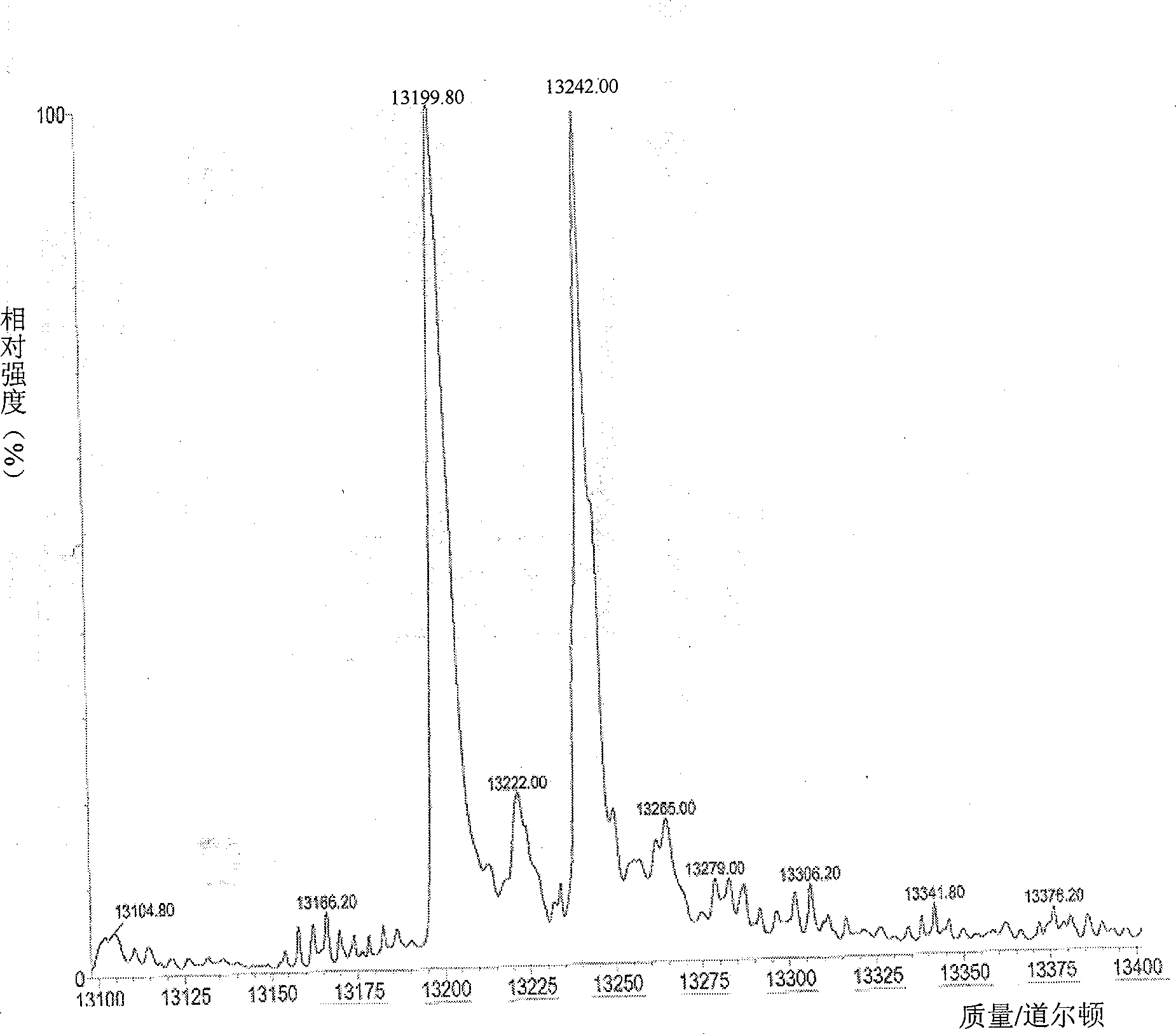
[0114] There are three known N-terminal acetyltransferases in Escherichia coli, namely RimI, RimJ, and RimL. There are also many genes of unknown function in the E. coli genome, among which there may also be unknown N-terminal acetyltransferases. The inventors selected five genes (yjaB, yjhQ, yjgM, yhhY, yiiD) presumed to be acetyltransferases from the genome. These 8 genes were knocked out by Red recombination technology, and different enzyme gene deletion mutants were constructed, in which Tα1 was expressed, separated and purified, and acetylated modified forms were identified by mass spectrometry. When the rimJ gene was knocked out, it was found that the fusion protein A2 had no acetylation modification, and it was deduced that this enzyme might be involved in the acetylation modification of Tα1. Now, the related experiments of rimJ gene knockout are described in detail as follows:
[0115]...
Embodiment 3
[0160] Example 3, Preparation of Thymosin Tα1 modified by N-terminal acetylation
[0161] The aforementioned studies have confirmed that rimJ is the key enzyme for the acetylation modification of Tα1. By co-expressing the Tα1 fusion protein with the rimJ gene, all the Tα1 fusion protein can be acetylated. In order to obtain the complete structure of Tα1, the fusion protein needs to be cleaved. The cleavage method can adopt enzymatic cleavage method, chemical cleavage method, and intein-mediated cleavage method. This example describes the use of intein cleavage to obtain N-terminal acetylated thymosin Tα1.
[0162] 1. Construction of engineering bacteria
[0163] (1) Construction of Tα1 and Spl DnaX intein fusion protein expression vector pET-AS
[0164] The gene sequence encoding the Spl DnaX intein is shown as sequence 9 in the sequence table, which was synthesized by Shanghai Sangon Bioengineering Technology Service Co., Ltd. Use restriction endonucleases EcoR I and Xho ...
PUM
| Property | Measurement | Unit |
|---|---|---|
| molecular weight | aaaaa | aaaaa |
| molecular weight | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com