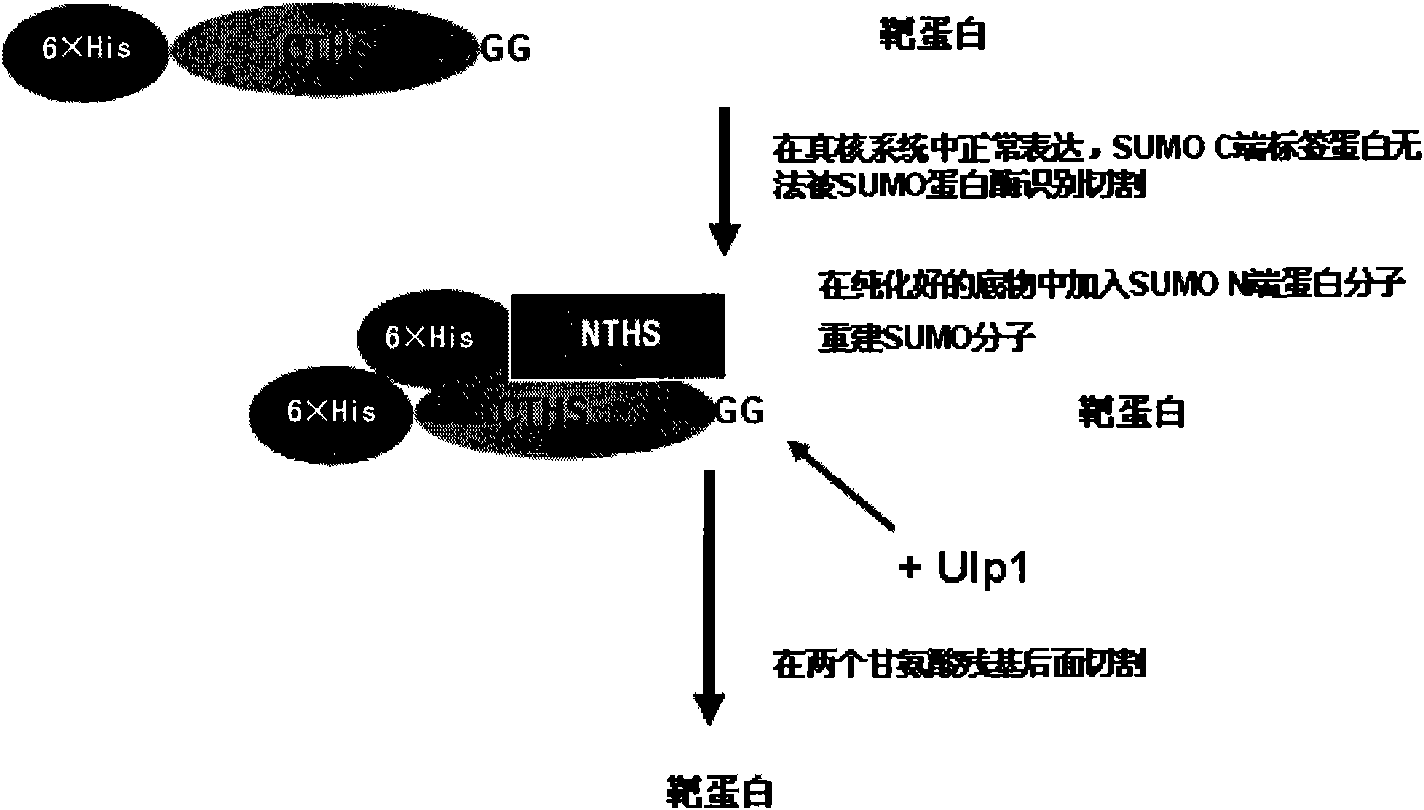
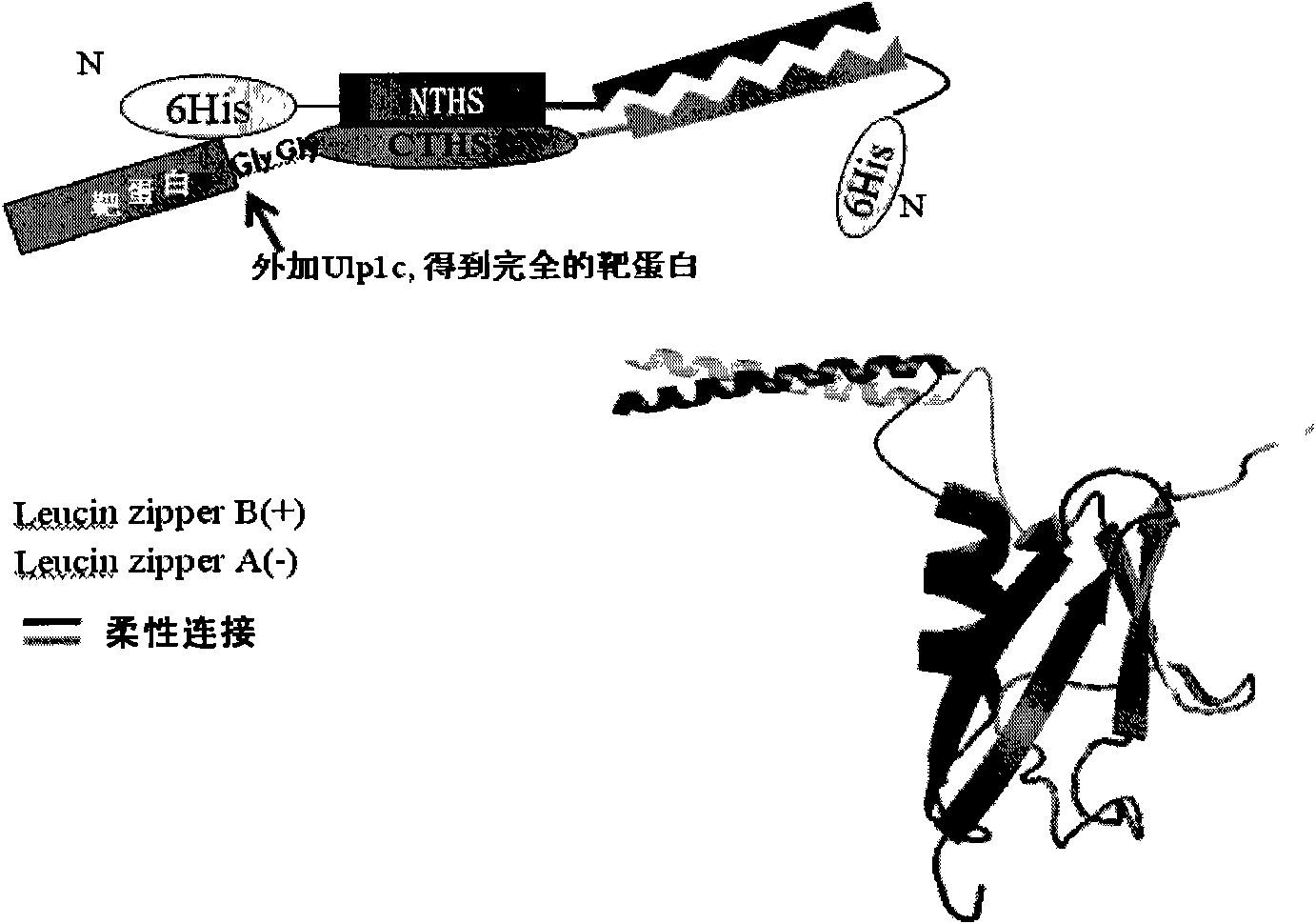
New-type label protein and application thereof
A protein and ubiquitin-like technology, applied in the direction of using vectors to introduce foreign genetic material, peptides, chemical instruments and methods, etc., can solve problems such as low cutting efficiency
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0132] Preparation of RbCl Competent Cells
[0133] (1) Streak Escherichia coli strains onto LB plates and incubate at 37°C for 10-12 hours.
[0134] (2) Pick a single clone in 30ml S.O.B (1:100 before adding 1M MgCl 2 , 1:100 by adding 1M MgSO 4 ), cultivate overnight at 37°C.
[0135] (3) 1:100 overnight bacteria into 200ml S.O.B (add 1M MgCl 1:100 before use) 2 , 1:100 added to 1M MgSO4), cultivated at 37°C until OD600=0.35.
[0136] (4) Quickly transfer the bacterial solution to an ice-salt bath (-3 to -5°C) and pre-cool for 15 minutes.
[0137] (5) Centrifuge at 4°C, 4500rpm×5min, and discard the supernatant. Add 64ml Competent Cell Buffer 1, suspend, and ice-salt bath for 15 minutes.
[0138] (6) Centrifuge at 4000rpm×5min at 4°C and discard the supernatant. Add 16ml Competent Cell Buffer 2 and suspend the cells on ice.
[0139] (7) Immediately aliquot into 200 μl / tube (tube pre-cooled).
[0140] (8) Quick-frozen in liquid nitrogen and stored at -70°C.
[0141]...
Embodiment 1
[0199] Example 1. Prokaryotic expression vector construction
[0200] The DNA sequences of SUMO, SUMO-CTHS, and Zipper-CTHS were inserted into pET28a through NcoI and XhoI sites respectively, and then the green fluorescent protein EGFP was inserted through EcoRI and XhoI as the target protein of the expression test, and the prokaryotic expression plasmid pET28a was constructed. - 6His-SUMO-EGFP, pET28a-6His-CTHS-EGFP, pET28a-6His-Zipper-CTHS-EGFP. And the 6His tag is fused at the N-terminus, which is convenient for purification and detection.
[0201] Construction of pET28a-6His-EGFP: Using pEGFP-N (Clontech Company) as a template, using primer1 (SEQ ID NO: 6) and primer2 (SEQ ID NO: 7) as primers, amplifying the EGFP gene, using EcoRI and XhoI After digestion, it was inserted into the pET28a vector (with 6His tag) that had undergone the same digestion.
[0202] Construction of pET28a-6His-SUMO-EGFP (SE): Using the Saccharomyces cerevisiae genome as a template, using primer3...
Embodiment 2
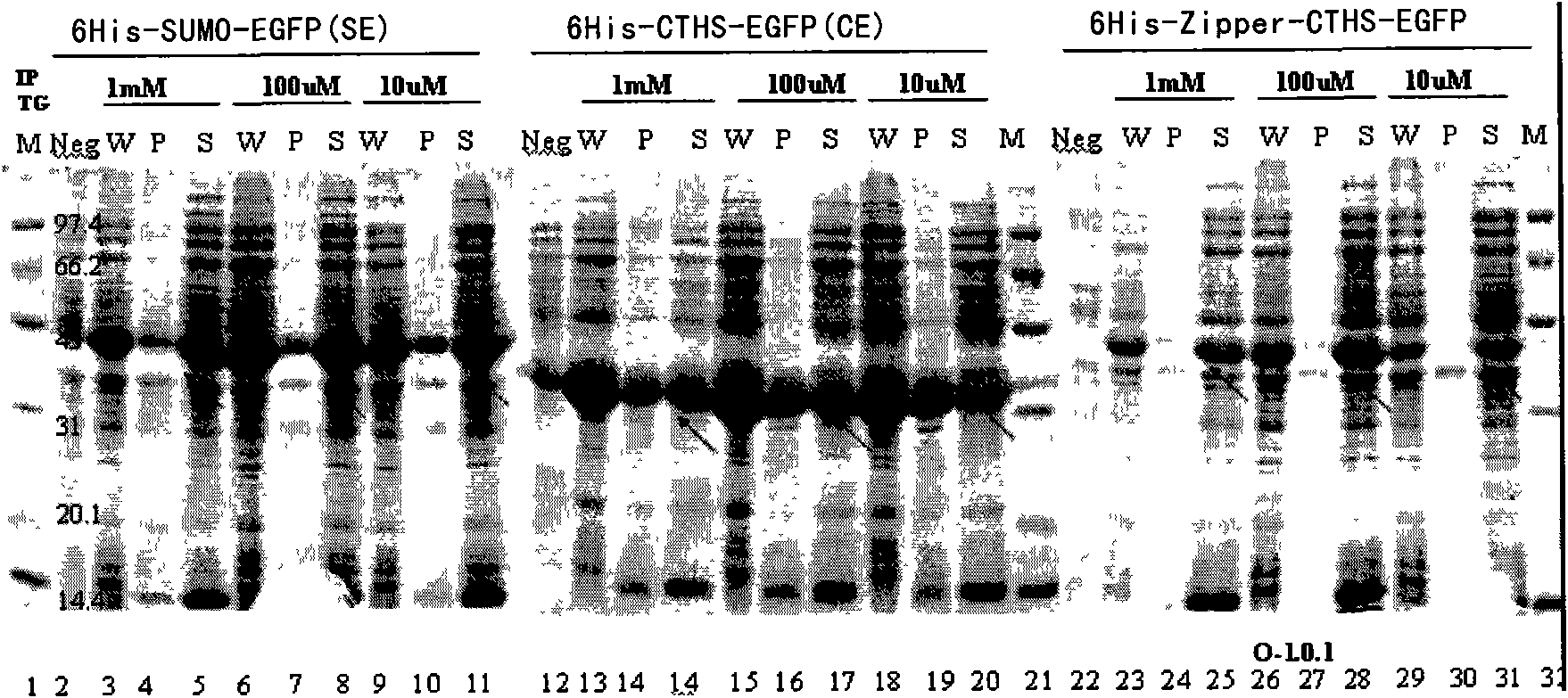
[0208] Example 2. Prokaryotic expression and solubility test
[0209] In this example, it is proved that in the E. coli expression system, Zipper (Zip)-CTHS has a good effect of promoting fusion protein solubility and enhancing fusion protein expression.
[0210] Expression of the fusion protein in E.coli: Transform the recombinant expression plasmids pET28a-6His-SUMO-EGFP, pET28a-6His-CTHS-EGFP, and pET28a-6His-Zipper-CTHS-EGFP into Escherichia coli (BL21 DE3 strain) respectively, and inoculate Monoclonal colonies were cultured in LB culture medium at 37°C for 12-14 hours; the above-mentioned culture liquid was inoculated into fresh LB medium at a ratio of 1:100, and cultured at 37°C until OD600 If it is 0.6-0.8, add isopropylthiogalactoside (IPTG) and cultivate at 22°C for 16-20h. The bacteria were collected by centrifugation at 6000rpm. Dissolve the bacteria in the bacteriostasis buffer containing 0.5mg / ml lysozyme, freeze and thaw with liquid nitrogen, repeat 3 times, add...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com