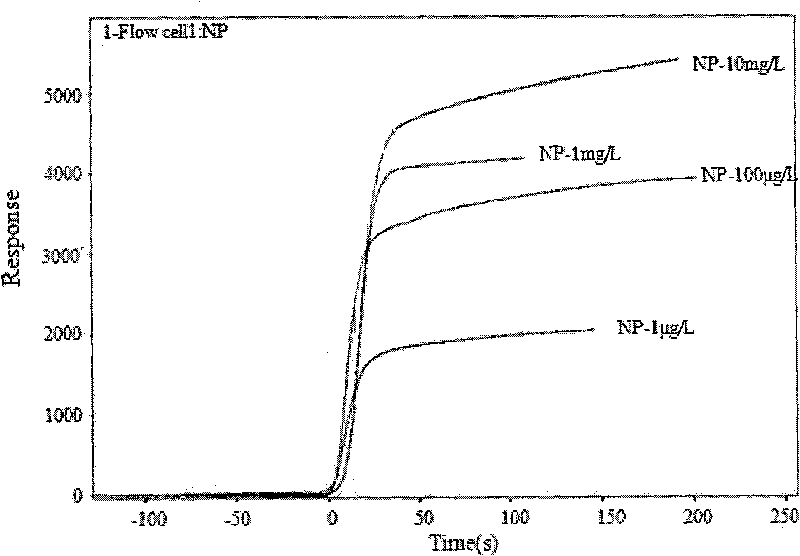
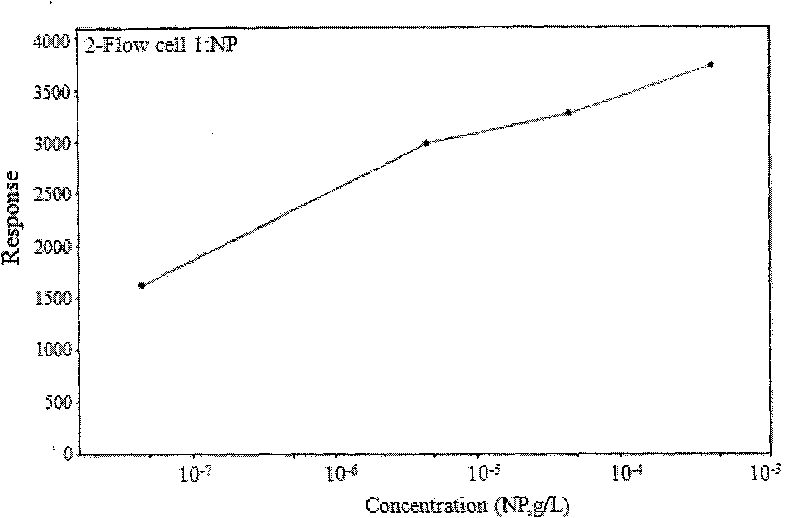
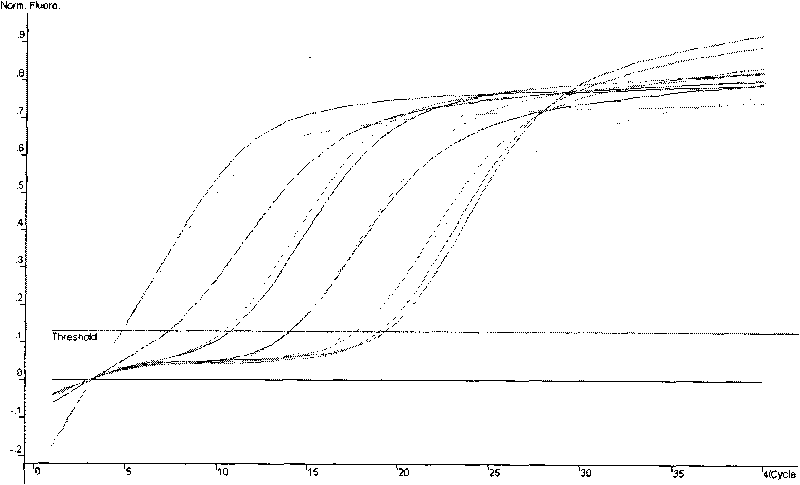
Method for detecting nonyl phenol by exonclease protection fluorescent quantitative PCR
A fluorescence quantification and nonylphenol technology, which is applied in the direction of fluorescence/phosphorescence, microbial measurement/inspection, biochemical equipment and methods, etc., can solve the problems of low sensitivity and high false positive rate, and achieve high sensitivity and detection limit Low, the effect of improving accuracy and sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0027] 1. Extraction of nonylphenol receptor and preparation of receptor-ligand complex
[0028] 1. Extraction of cytosol containing nonylphenol receptors
[0029] Goldfish with similar body length and weight were first sterilized with 5% saline solution, and then domesticated in the laboratory for a week with tap water that had been naturally dechlorinated for 3 days. During the domestication process, feed once a day and aerate with a sand head. One week later, nonylphenol was added to make the concentration of nonylphenol in the water 0.01mg / L. During the feeding process, the static fluid replacement method was adopted and replaced every 24 hours. After one month of domestication, the goldfish were taken out and anesthetized with 5 mg / L benzocaine. Take the liver, wash off the blood outside the liver with 0.15mol / L KCl solution pre-cooled on ice, then blot the surface water with sterilized filter paper, and add HEDG buffer (pre-cooled on ice) was homogenized in a glass ho...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com