Method for analyzing and detecting pathogenic microorganisms
A pathogenic microorganism and bioluminescence technology, applied in the field of micro-total analysis technology in the field of biological analysis, can solve the problems of strict culture environment and operator requirements, unsuitable rapid detection of pathogenic bacteria, accumulation of operating errors, etc., to ensure the detection range and avoid samples. loss, the effect of reducing operating errors
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
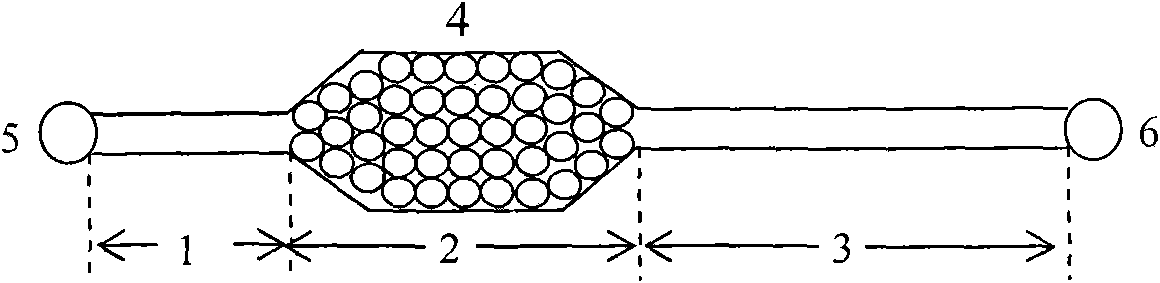
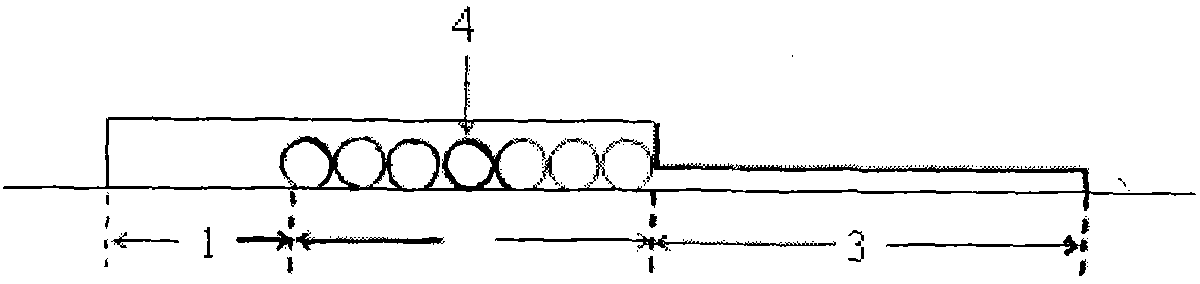
[0038] Example 1 :use figure 1 Chips shown for E.coli O157:H7 capture / enrichment studies
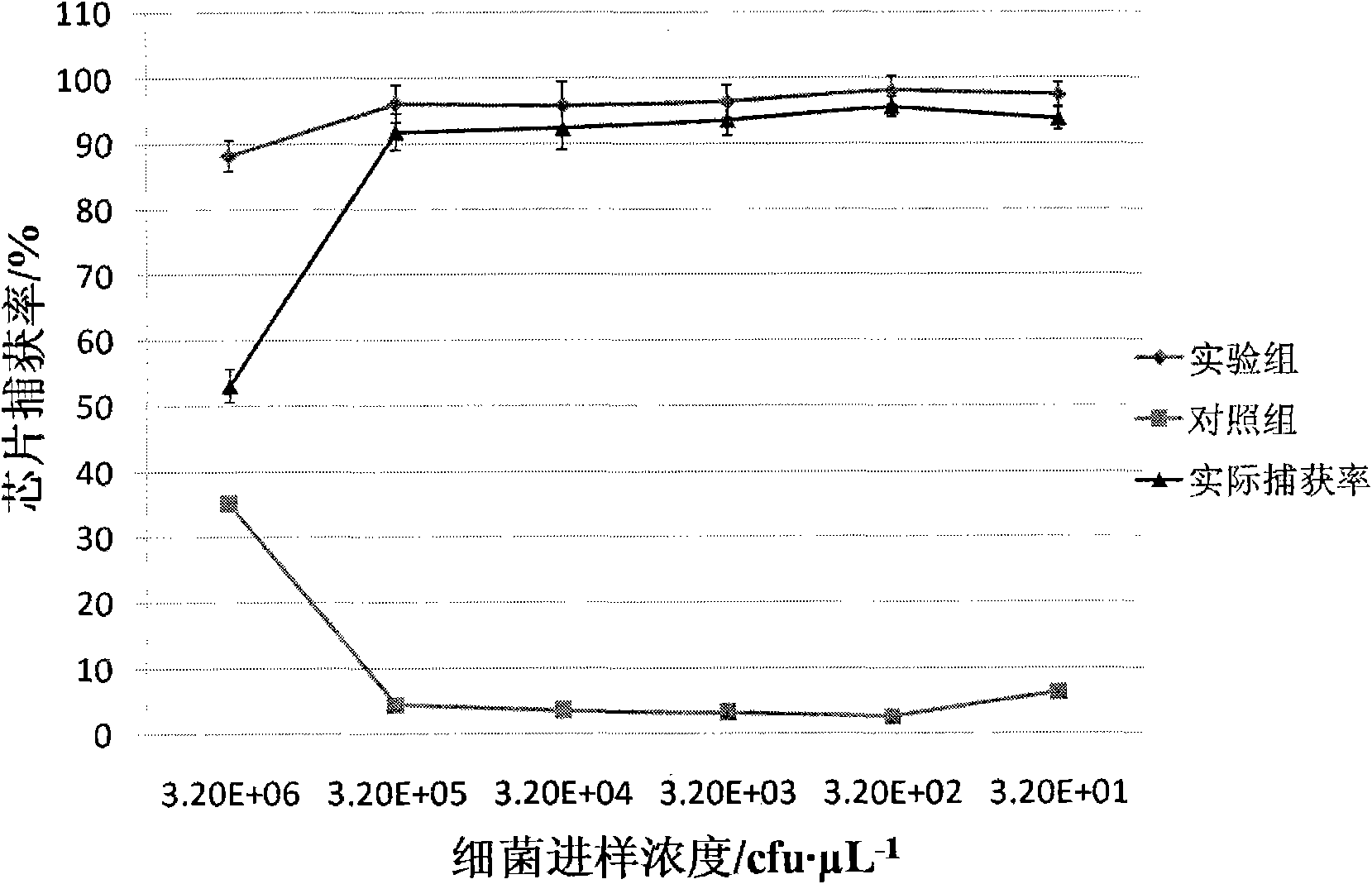
[0039] Use 0.01mol L -1 PBS Press 10, 10 2 , 10 3 , 10 4 , 10 5 After fully suspended, take 1 μL of the above-mentioned bacterial suspension (the concentration of the bacterial solution is shown in Table 1) and add it to the inlet storage tank, and the concentration is 5 μL·min. -1 The flow rate is driven by positive pressure into the microchannel of the chip, and then 0.01mol L with pH=7.20 -1 PBS at 10 μL·min -1 Flush the microchannel for 5 min at a flow rate. In the whole study, the microbead chip filled with antibody modification was used as the experimental group, and the microbead chip filled with BSA blocked as the control group. Using the plate counting method, culture and count the bacterial sample injection liquid and the flushing liquid collected in the outlet reservoir, according to the formula:
[0040]
[0041] Calculate the capture rate and the adsorption rat...
Embodiment 2
[0046] Example 2 : Bioluminescence detection of standard strain E.coli O157:H7
[0047] Use 0.01mol L -1 Diluted 10 times with PBS buffer, the bacterial concentration was 10 5 cfu μL -1 . Take 1 μL of the bacterial suspension and add it to the inlet reservoir of the chip. -1 The flow rate is driven into the chip by positive pressure, and then 0.01mol L with pH=7.20 -1 PBS buffer at 10 μL min -1 Flush at a flow rate of 5 minutes to reduce or eliminate non-specific adsorption or residues on the chip wall and in the gap between microbeads. The bioluminescence reagent (BacTiter-Glo TM Microbial Cell Viability Assay, Promega, USA) at 1 μL min -1 The positive pressure of the flow rate is transmitted to the chip, and the luminescence intensity is detected at the chip outlet reservoir as the luminescence detection point, and the signal is collected by the PMT optical signal detector, and the typical luminescence curve is as follows: image 3 shown.
[0048] According to th...
Embodiment 3
[0051] Example 3: Bioluminescent detection of E.coli O157:H7 in food samples
[0052] Take two portions of clean pork, 5g each, cut into 1mm in a sterile environment 3 Left and right pieces. Add two portions of pork to two sterilized centrifuge tubes, then add 1000 μL of E.coliO157:H7 that has been proliferated for 8 hours and dilute 10 3 times E.coli O157:H7 1000μL each, and 0.01mol·L -1 PBS 4mL each one. After the above two groups of pork samples containing different concentrations of bacterial suspension were vortexed and mixed, they were centrifuged at 1000 rpm for 5 minutes, and then 1 μL of supernatant (see Table 3 for the concentration of the injected bacterial solution) was added to the chip inlet reservoir. , at 5 μL·min -1 The flow rate is driven into the chip by positive pressure, and then 0.01mol L with pH=7.20 -1 PBS buffer at 10 μL min -1 Rinse at flow rate for 5 minutes. The bioluminescent reagent was added at 1 μL min -1 The positive pressure of the flow ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com