Method for detecting Chikungunya virus nucleic acid by real-time fluorescent quantitative PCR
A technology for real-time fluorescence quantification and viral nucleic acid, applied in the fields of fluorescence/phosphorescence, microbial measurement/inspection, biochemical equipment and methods, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0030] Example 1: Detection of Primer Sensitivity
[0031] 1) Select a highly conserved region fragment in the whole genome of the virus as a standard, and its viral RNA from 2.3 × 10 10 copy / uL was sequentially diluted to 2.3×10 -1 copy / uL, select 2.3×10 6 copy / uL to 2.3×10 -1 The copy / uL copy number gradient is used for detection, the experiment is repeated 3 times, and the average value is taken;
[0032] 2) Negative control: nuclease-free water;
[0033] 3) The reaction system is prepared according to Table 2;
[0034] 4) At the same time, it is compared with the currently commercially available chikungunya virus nucleic acid detection kit.
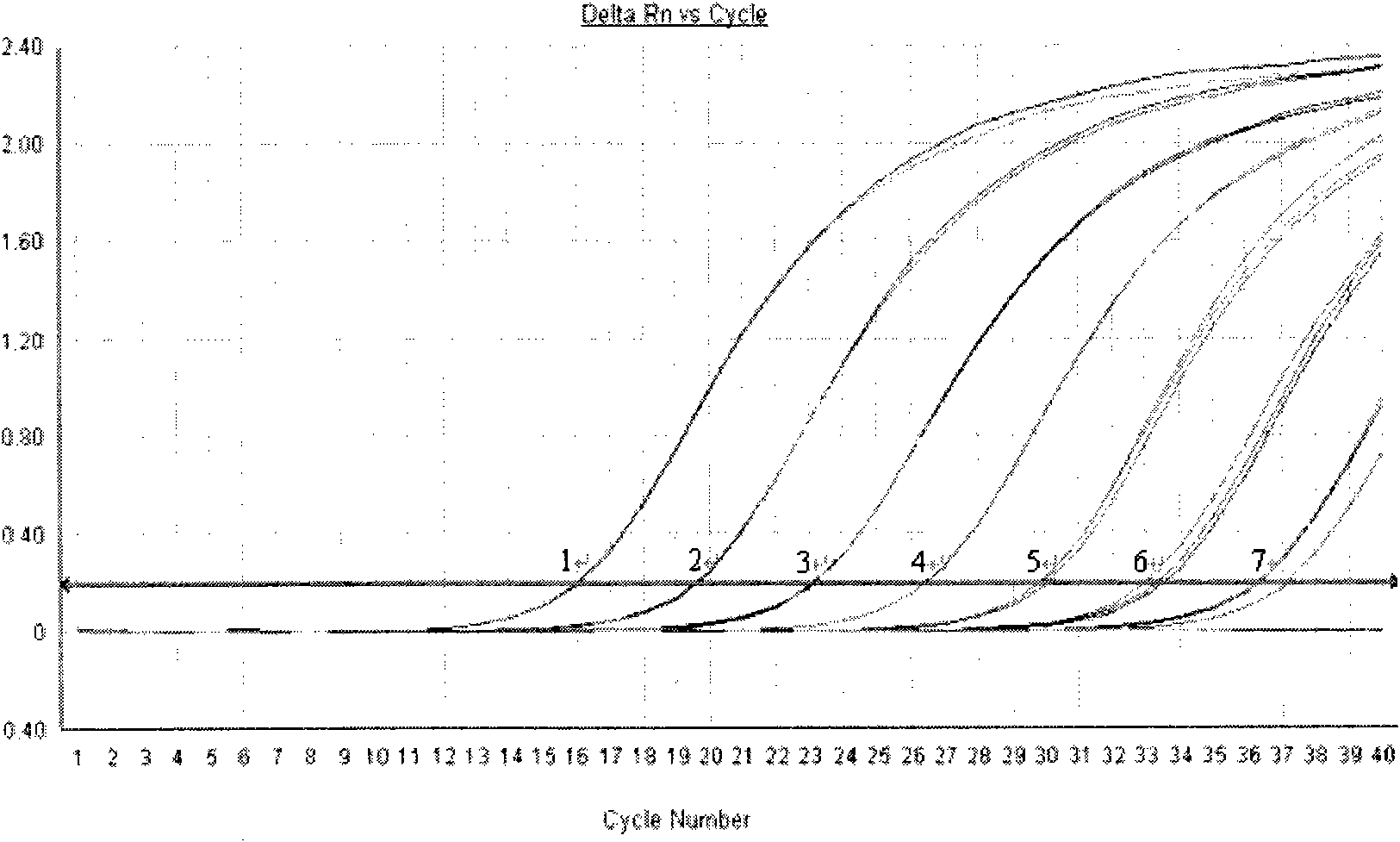
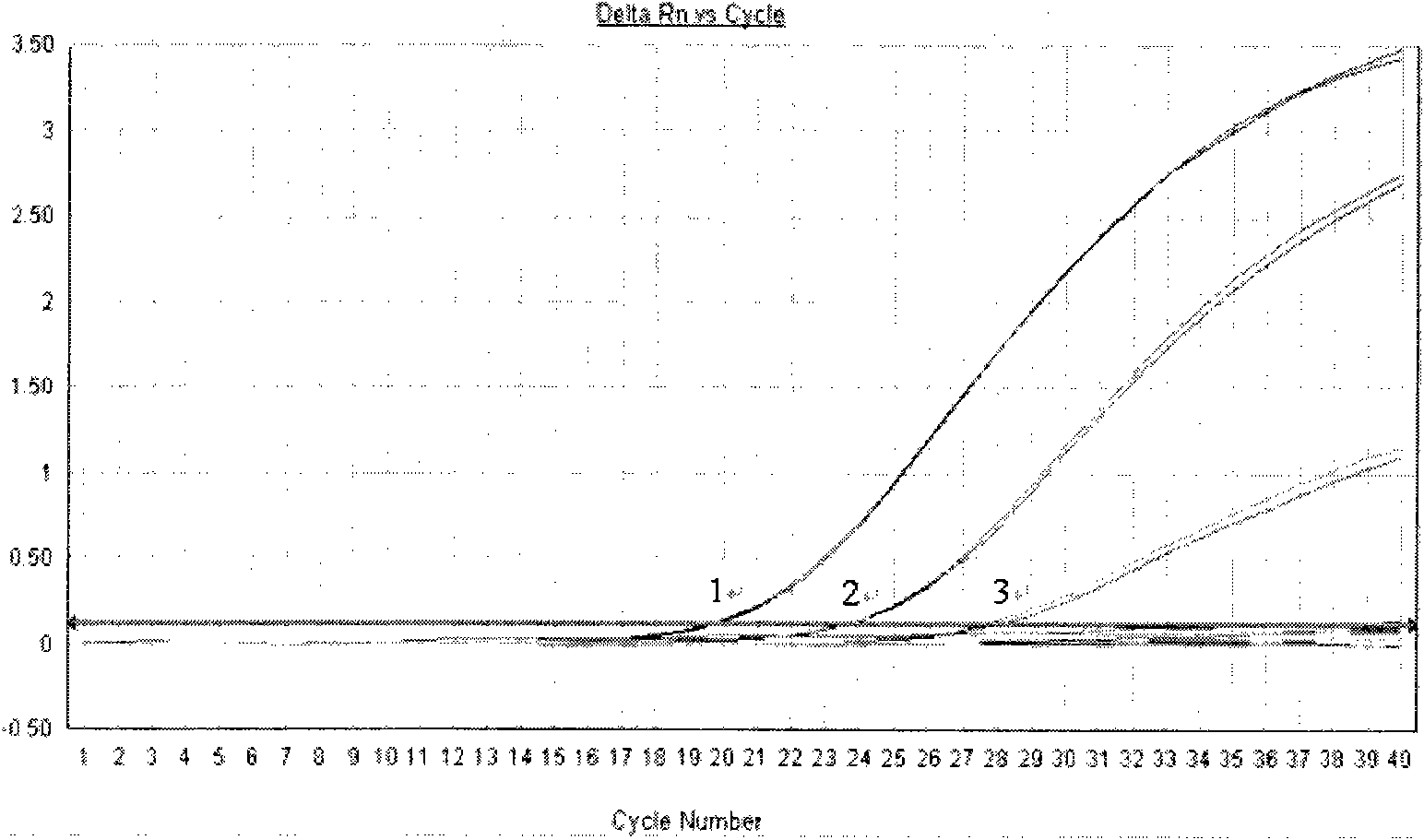
[0035] Test results such as figure 2 , image 3 , Table 3, Table 4 shown.
[0036] figure 2 Among them, the curves 1-7 are respectively the respective dilutions of the standard substance in the present invention 2.3×10 6 copy / uL-2.3×10 0 The curve of copy / uL amplification.
[0037] image 3 Among them, the curves 1-3 r...
Embodiment 3
[0044] Embodiment 3: Primer detection unknown sample
[0045] 1) Detect the collected unknown samples, screen the Chikungunya virus and monitor the non-specificity of the primers;
[0046] 2) Positive control: virus standard (2.3×10 3 copy / uL);
[0047] 3) The reaction system was prepared according to Table 2.
[0048] See the test results Figure 4 and Table 5.
[0049] Figure 4Only one amplification curve appeared in the positive control, and the rest of the samples had no amplification. The reaction was negative. PTC was used as a positive control.
[0050] The data in Table 5 shows that the results of testing unknown samples were all negative.
[0051] Table 6
[0052]
[0053] Result analysis:
[0054] After the amplification reaction completed 40 cycles, the minimum detection limit of the primer and probe detection positive standard in the present invention was 2.3copy / uL, while the minimum detection limit of the currently commercially available chikungunya...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com