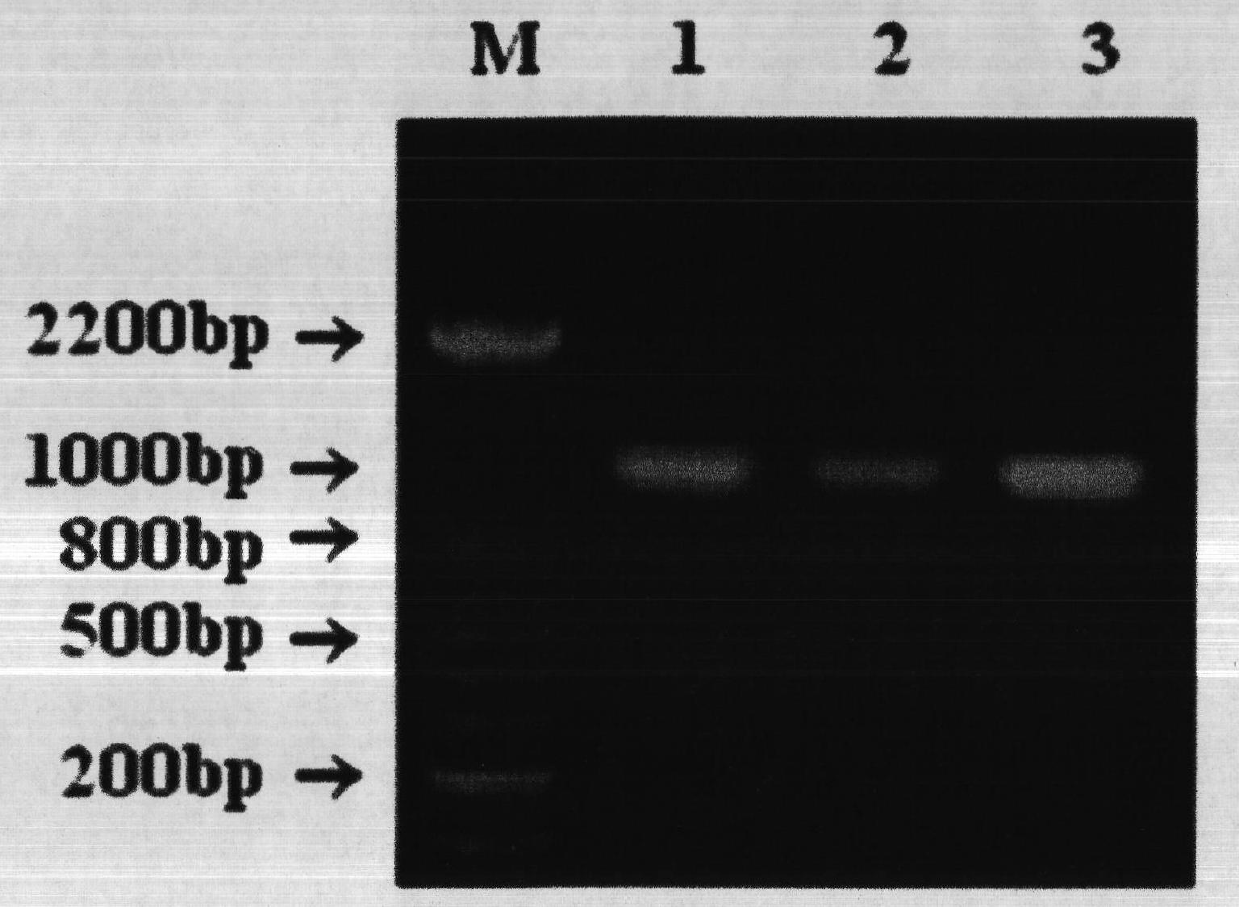
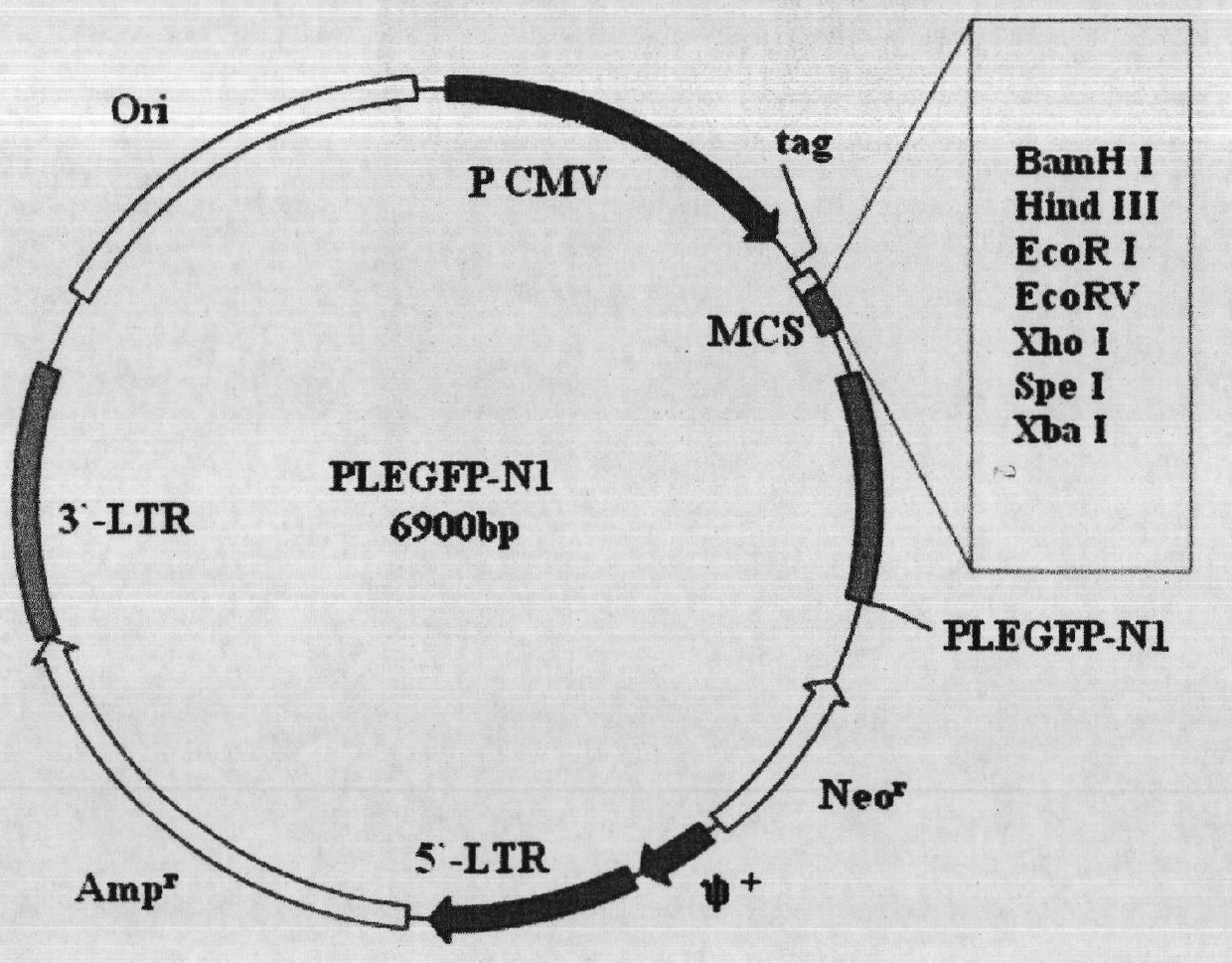
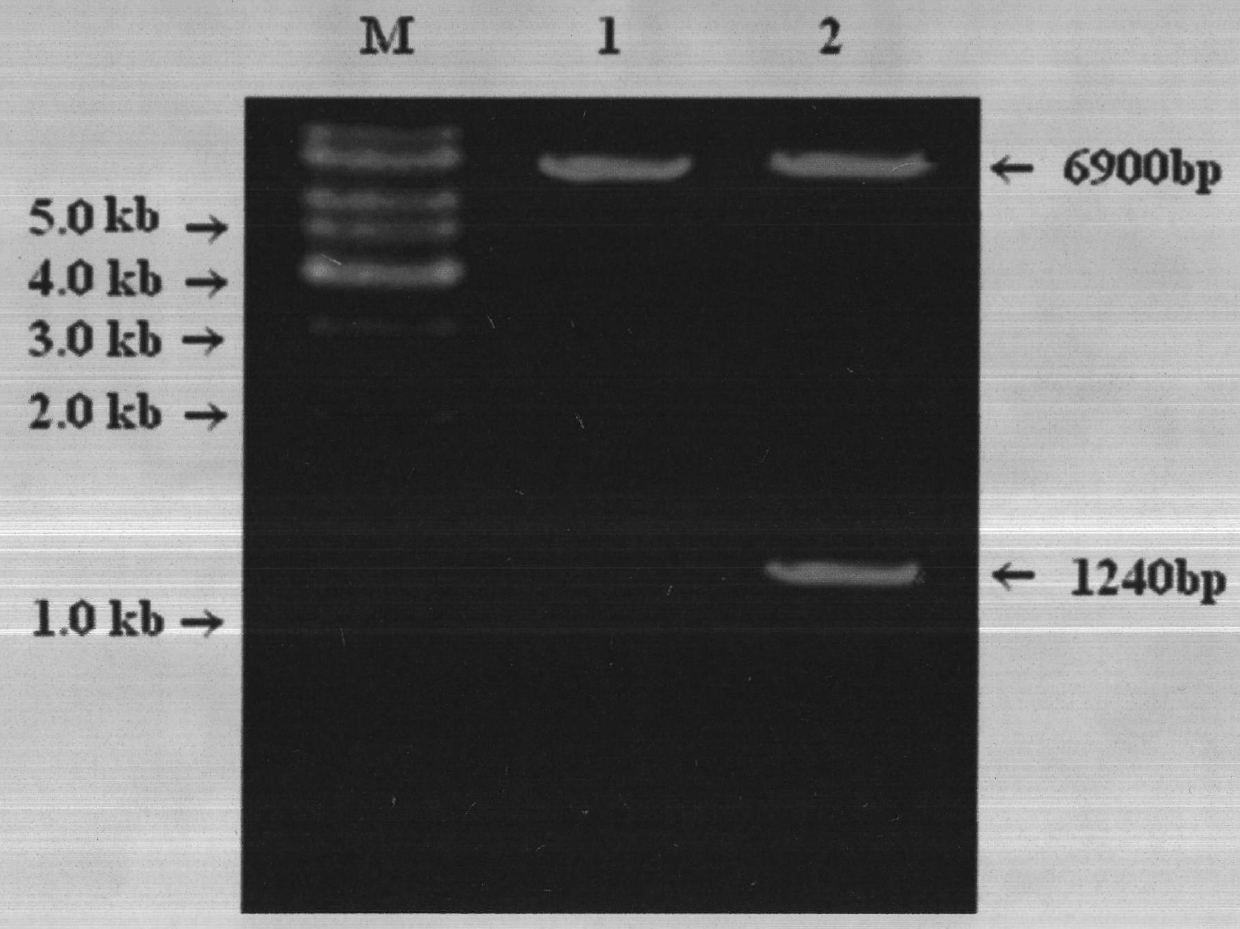
Construction and application method of efficient double promoter PLEGFP-N1-spMyoD1 green fluorescence retrovirus vector
A technology of PLEGFP-N1, plegfp-n1-spmyod1, applied in the directions of botanical equipment and methods, biochemical equipment and methods, introduction of foreign genetic material using vectors, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
preparation example Construction
[0109] 2.6 Preparation of Competent Cells:
[0110] Apply CaCl 2 Prepare Escherichia coli competent DH5a strains by sensitization method. After overnight shaking culture at 37°C and 225rpm, the bacterial liquid was streaked on LB solid medium, cultured overnight in a 37°C incubator, and single clones were screened. Select a single colony and inoculate in 5mL LB medium, culture overnight, inoculate the culture in LB medium at 1 / 100, and cultivate until A 600 Up to 0.45-0.55, with CaCl 2 Sensitized Escherichia coli. The details are as follows: after 10 minutes of ice-bathing the bacterial solution, centrifuge at 4000 rpm for 20 minutes at 4°C to collect the bacterial cells, and then add ice-cold 0.1M CaCl 2 Sensitization solution, suspended bacteria, ice-bathed for 10 minutes, after re-collecting the bacteria, add ice-cold 0.1M CaCl 2 , resuspend the cells, add glycerol to a final concentration of 15%, aliquot 100 μL / tube, store at -70°C, and use for transformation.
[0111...
Embodiment 1
[0218] Liposome 2000-mediated expression of PLEGFP-N1-spMyoD1 vector plasmid in C2C12 cells. C2C12 cells not transfected with expression vector, cultured under the same conditions, and with the same concentration were used as negative control.
[0219] method:
[0220] Take the cultured C2C12 cells when the concentration reaches 50%, and add 2ml of antibiotic-free DMEM+10% fetal bovine serum culture medium to the cells, and the cells grow to 80%-90%, and the transfection can begin. At the same time, 30ul of 20ng / ul vector plasmid was added to Opti-MEM 220ul serum-free medium, with a total volume of 250ul, mixed gently, and left for 5 minutes. Take liposome 200010ul, add to Opti-MEM 240ul serum-free medium, total volume 250ul, mix gently. Leave for 5 minutes. Add the liposome mixture to the vector plasmid mixture and mix gently; let stand for 20 minutes. Change the medium of the cells every 20 minutes, add 1.5ml of antibiotic-free medium (DMEM+10% fetal bovine serum), and a...
Embodiment 2
[0223] Liposome 2000-mediated expression of PLEGFP-N1-spMyoD1 vector plasmid in 293T cells, 293T cells not transfected with expression vector, cultured under the same conditions, and with the same concentration were used as negative control.
[0224] method:
[0225] Take the cultured 293T cells when the concentration reaches 50%, add 2ml of antibiotic-free DMEM+10% fetal bovine serum culture medium to the cells, and the cells grow to 80%-90%, and the transfection can start. At the same time, 30ul of 20ng / ul vector plasmid was added to Opti-MEM 220ul serum-free medium, with a total volume of 250ul, mixed gently, and left for 5 minutes. Take liposome 2000 10ul, add to Opti-MEM 240ul serum-free medium, total volume 250ul, mix gently. Leave for 5 minutes. Add the liposome mixture to the carrier plasmid mixture and mix gently; let stand for 20 minutes. Change the medium of the cells every 20 minutes, add 1.5ml of antibiotic-free medium (DMEM+10% fetal bovine serum), and add a ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com