microRNA biomarker and application thereof
A biomarker, mir-25 technology, applied in the field of microRNA biomarkers and its application in the detection of liver cancer patients, can solve the lack of systematic analysis reports on serum RNA stability research, and the limited storage time of miRNA stability research , temperature and repeated freezing and thawing
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0108] Example 1 Biomarkers and Detection Primers and Probes
[0109] The present invention provides a set of miRNA biomarkers comprising miR-21, miR-25, miR-29c, miR-93, miR-198, miR-221 and miR-222. The specific sequence of this group of biomarkers is as follows:
[0110] miR-21: uagcuuaucagacugauguuga (SEQ ID NO: 1)
[0111] miR-25: cauugcacuugucucggucuga (SEQ ID NO: 2)
[0112] miR-29c: uagcaccauuugaaaucgguua (SEQ ID NO: 3)
[0113] miR-93: caaagugcuguucgugcagguag (SEQ ID NO: 4)
[0114] miR-198: gguccagaggggagauagguuc (SEQ ID NO: 5)
[0115] miR-221: agcuacauugucugcuggguuuc (SEQ ID NO: 6)
[0116] miR-222: agcuacaucuggcuacugggu (SEQ ID NO: 7)
[0117] Stem-loop primer method: Using the stem-loop primer method to detect the expression of miRNA requires specific stem-loop primers and PCR primers for the specific sequence of the miRNA. During the PCR amplification process, SYBR-green color development was used to quantitatively detect the amplification of the target m...
Embodiment 2
[0142] Embodiment 2 Experimental material and experimental reagent
[0143] In the present invention, the liver cancer cell line Huh-7, liver cancer tissues, 22 clinical normal serum samples (including 11 males and 11 females) and 10 clinical liver cancer serum samples were used. All of the above are isolated tissues.
[0144] The experimental reagents are commonly used molecular biology reagents, mainly including: Trizol reagent, chloroform, isopropanol, ethanol, DNA polyacrylamide gel electrophoresis reagent and other conventional biochemical reagents were purchased from Shanghai Sangong Company. M-MLV reverse transcriptase, RNase inhibitor, and dNTP mixture for reverse transcription (each 10 mM, RNase-free) were purchased from TaKaRa Company. Fluorescence quantitative PCR kit is a TOYOBO product.
Embodiment 3
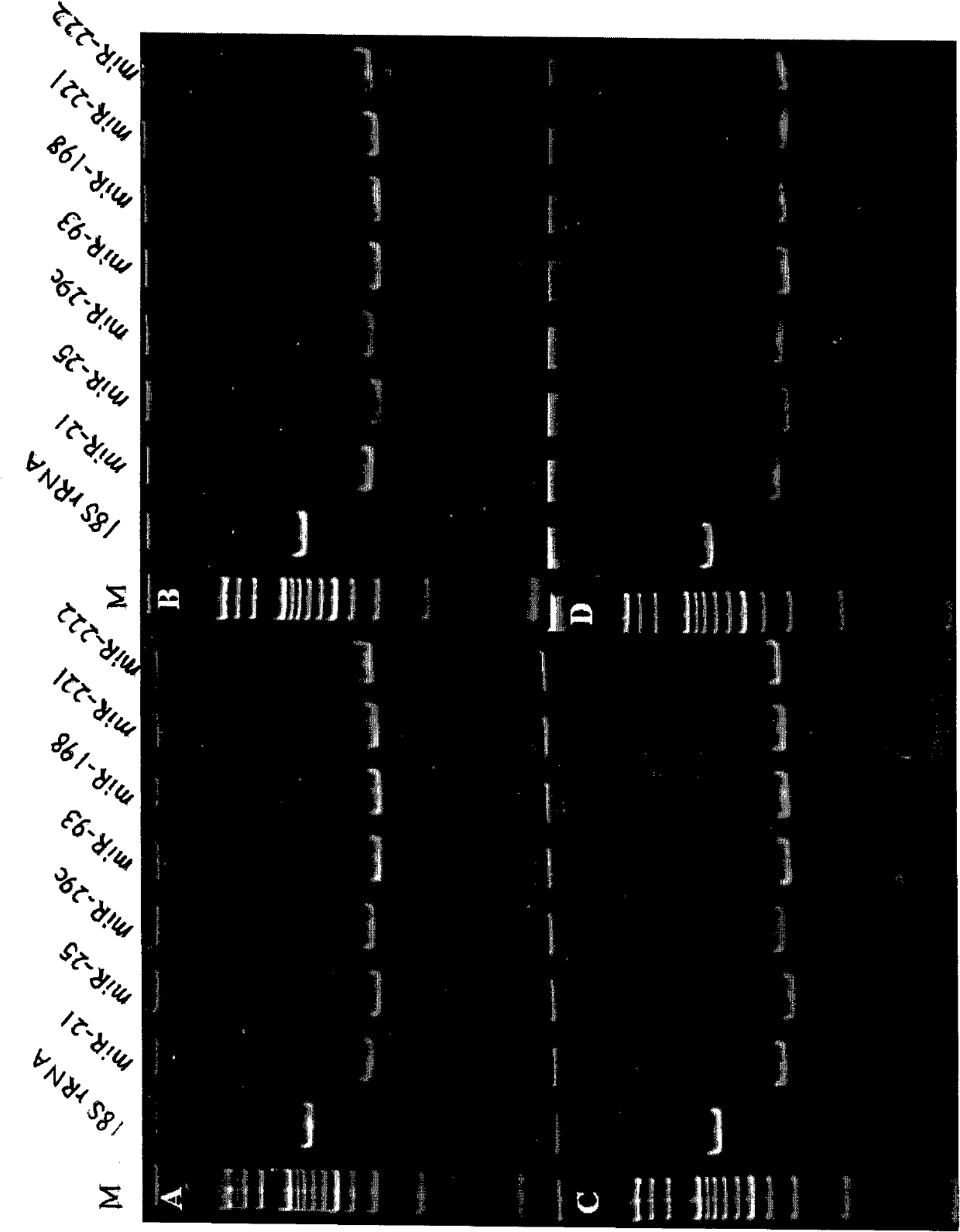
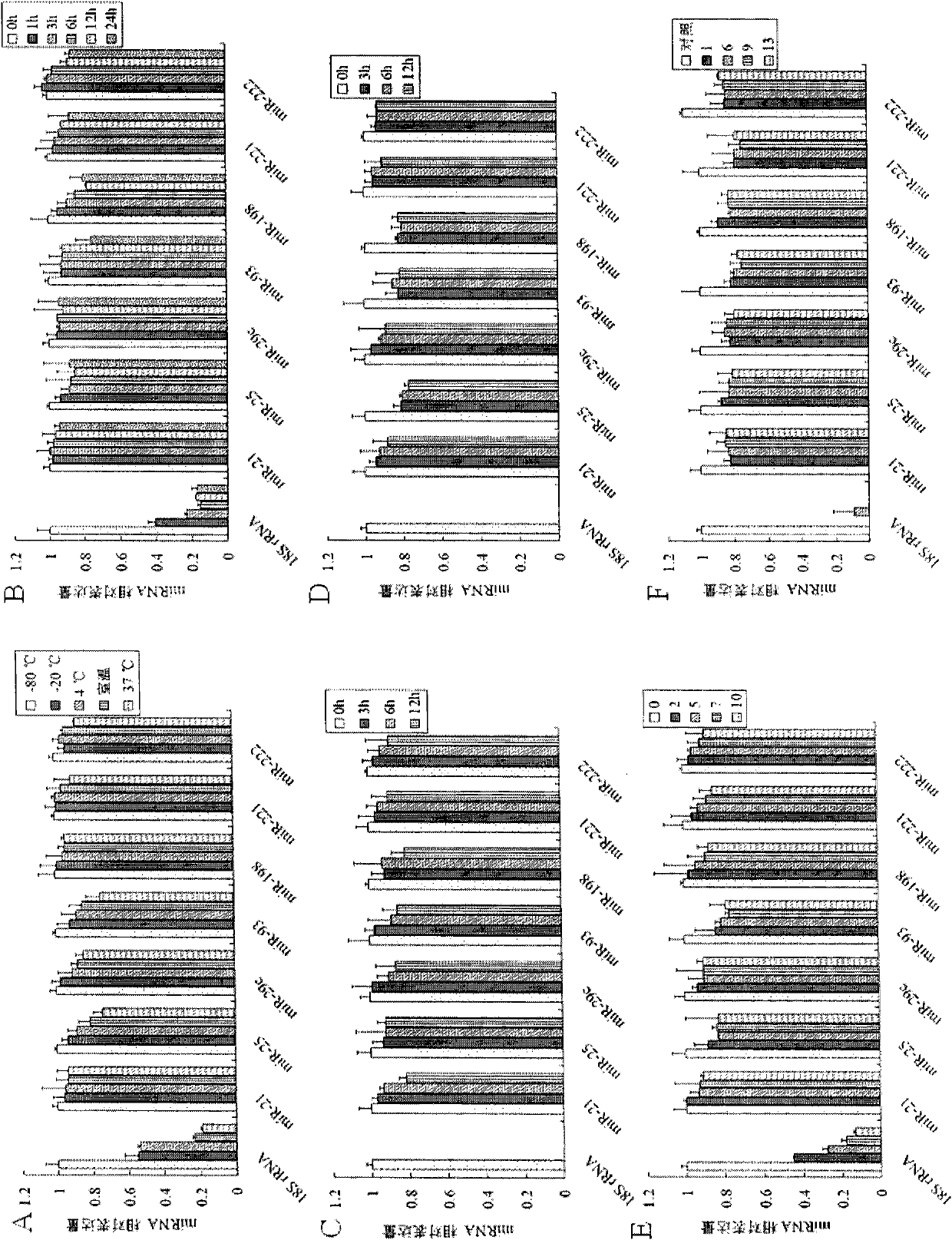
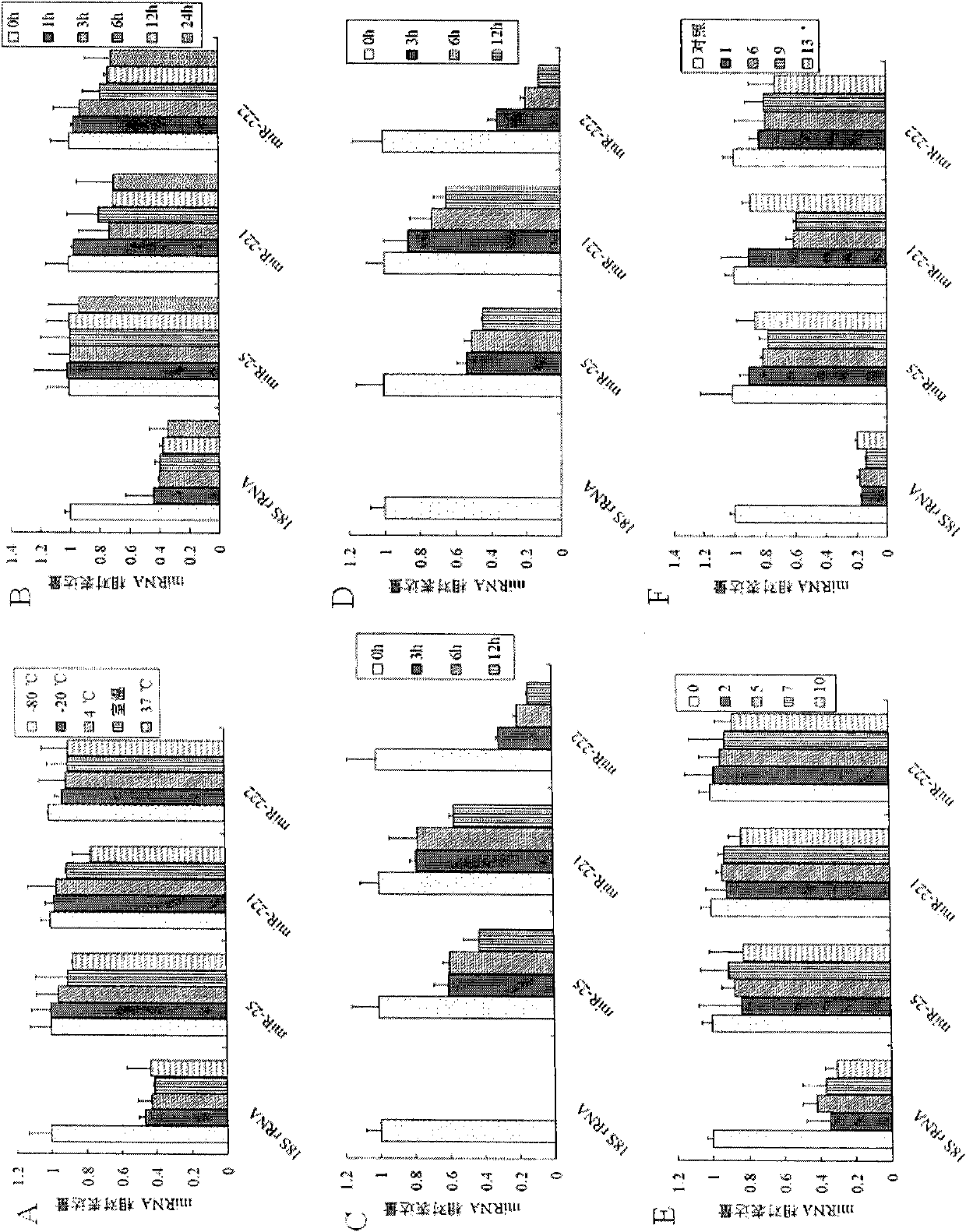
[0145] Example 3 Detection of miRNA expression in liver cancer cell lines, tissues and serum by stem-loop primer method
[0146] 1. cDNA synthesis
[0147]Add 100ng of total RNA of liver cancer Huh-7 cell line, 100ng of total RNA of liver cancer tissue, 40ng of RNA extracted from serum, and 13μL of serum to four RNase-free 0.2mL PCR tubes. If the total volume of the above is less than 13μL, add DEPC water to supplement. to a total volume of 13 μL, then denatured at 70°C for 8 min, and then quickly placed on ice for 3 min, then sequentially added 0.5 μL RNase inhibitor (40 U / μL), 1 μL dNTP mixture (each 10 mM, RNase-free), 4 μL 5×M-MLV buffer solution, 1 μL of the stem-loop primer (2 μM) and 0.5 μL of M-MLV reverse transcriptase (200 U / μL) as described in Example 1, to make the total volume to 20 μL, mix well and centrifuge slightly, and carry out the first step of cDNA in a PCR machine. For one-strand synthesis, the reaction parameters were set at 16°C for 15 minutes, 42°C fo...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com