Drug screening model of targeting coronavirus protease and application thereof
A coronavirus and protease technology, applied in the direction of microorganisms, microorganism-based methods, microorganism measurement/inspection, etc., can solve the problems of high risk for experimenters, virus leakage, inconvenient operation, etc., to avoid virus infection and dynamic screening Effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment Construction
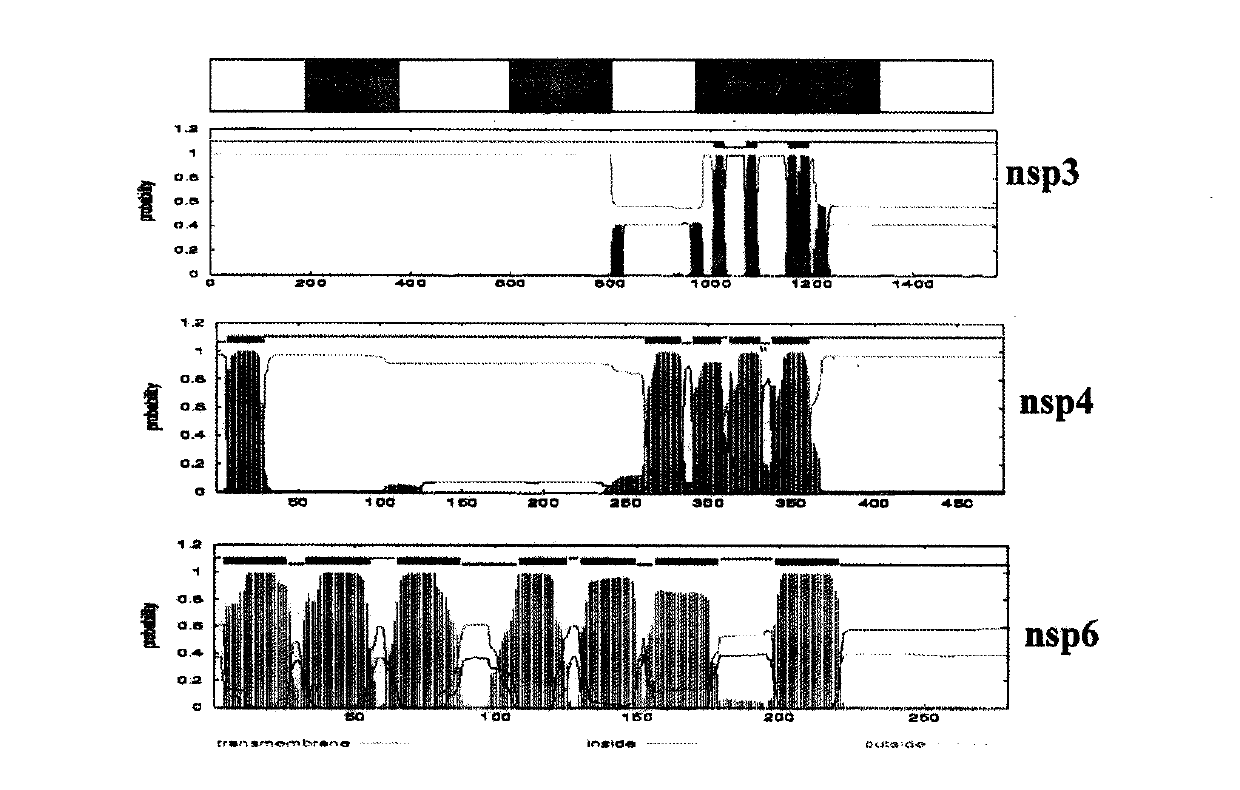
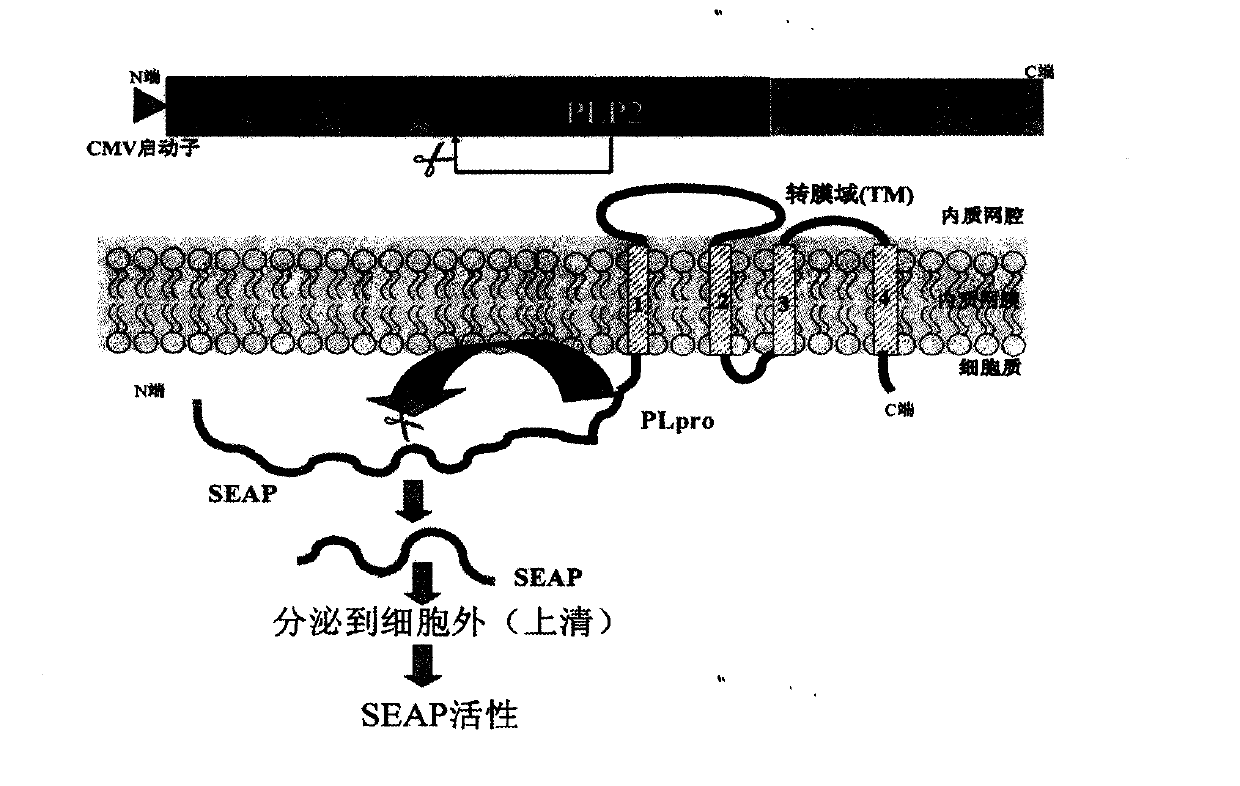
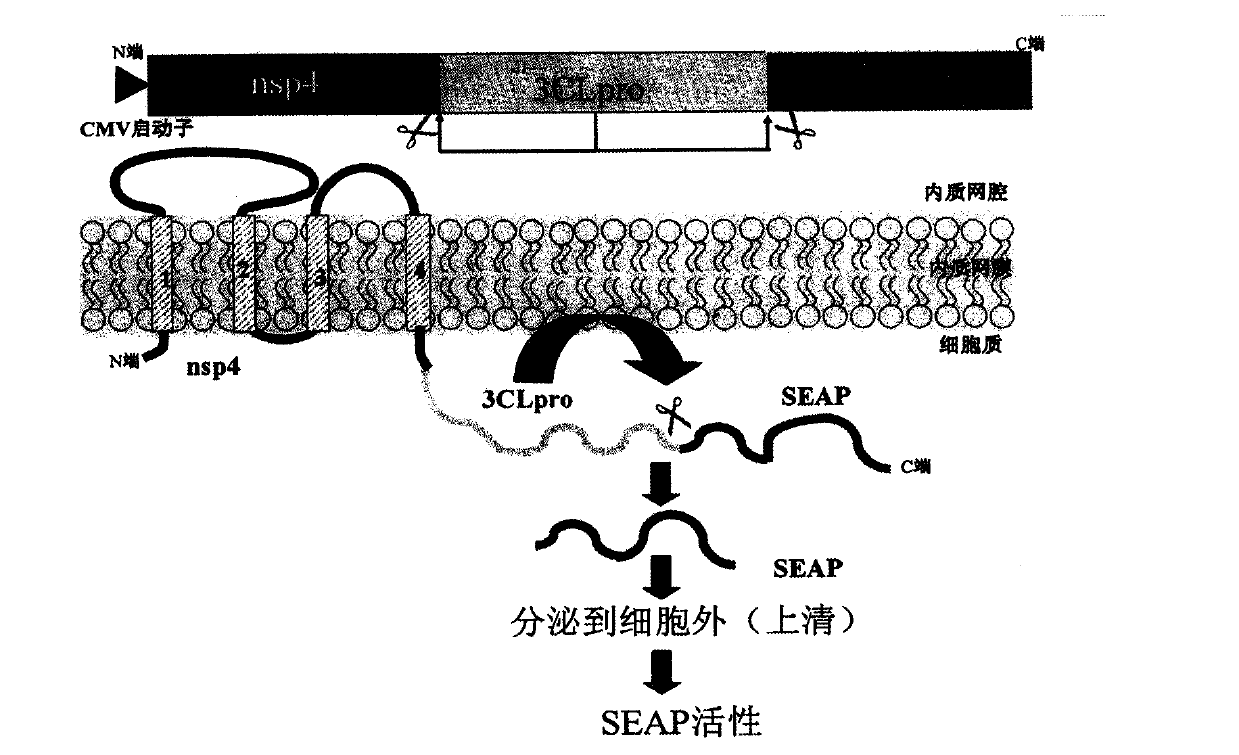
[0019] 1. Drug screening model targeting coronavirus PLP protease: clone the PLP2-TM fragment of NL63 into the vector pcDNA3.1-V5 / His B (Xho I and Apa I). The amino acid sequence of the nsp2 / 3 site recognized by PLP2 -FTKLAG↓GK- is carried on the N segment of PLP2. Using SEAP-basic vector (Clontech) as a template, PCR obtained the SEAP fragment (with EcoR I and Kozak sequences and SEAP ATG at the 5' end, and XhoI at the 3' end), and cloned it into pcDNA-V5 / HisB PLP2-TM At the 5' end of the fragment, a 5'-SEAP-PLP2-TM-3' DNA construct (pcDNA-SEAP-PLP2-TM) was obtained. The recombinant DNA plasmid was transfected into Hela cells, and 50 μl of supernatant was collected at each time point of 12h, 24h, 36h, 48h and 60h after transfection. The SEAP activity in the supernatant was detected by Chemiluminescent Assay (the reagent was Great EscAPe SEAP Chemiluminescent and Fluorescence Detection Kits, Clontech Company).
[0020] In order to confirm that PLP2 can release SEAP into the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com