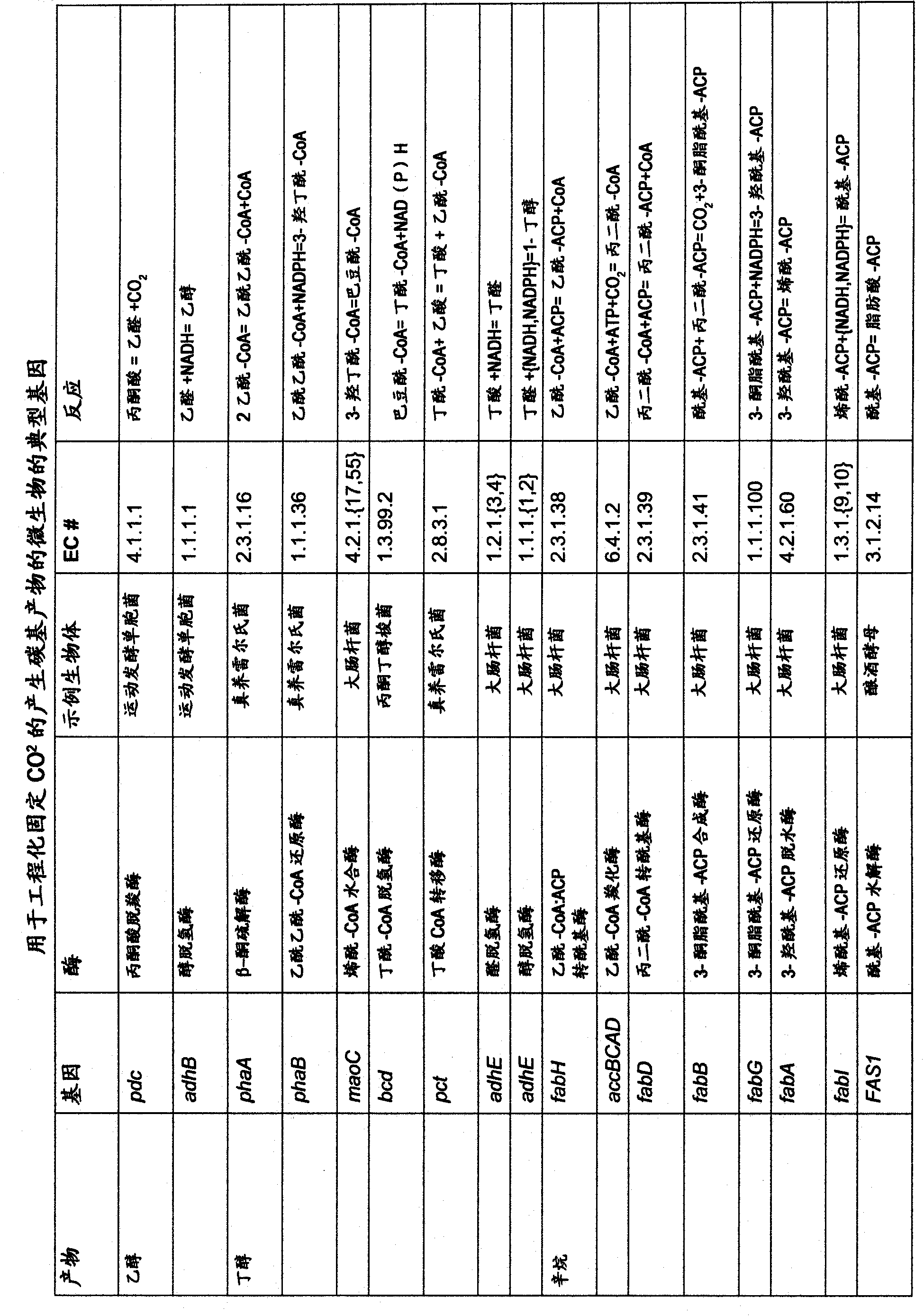
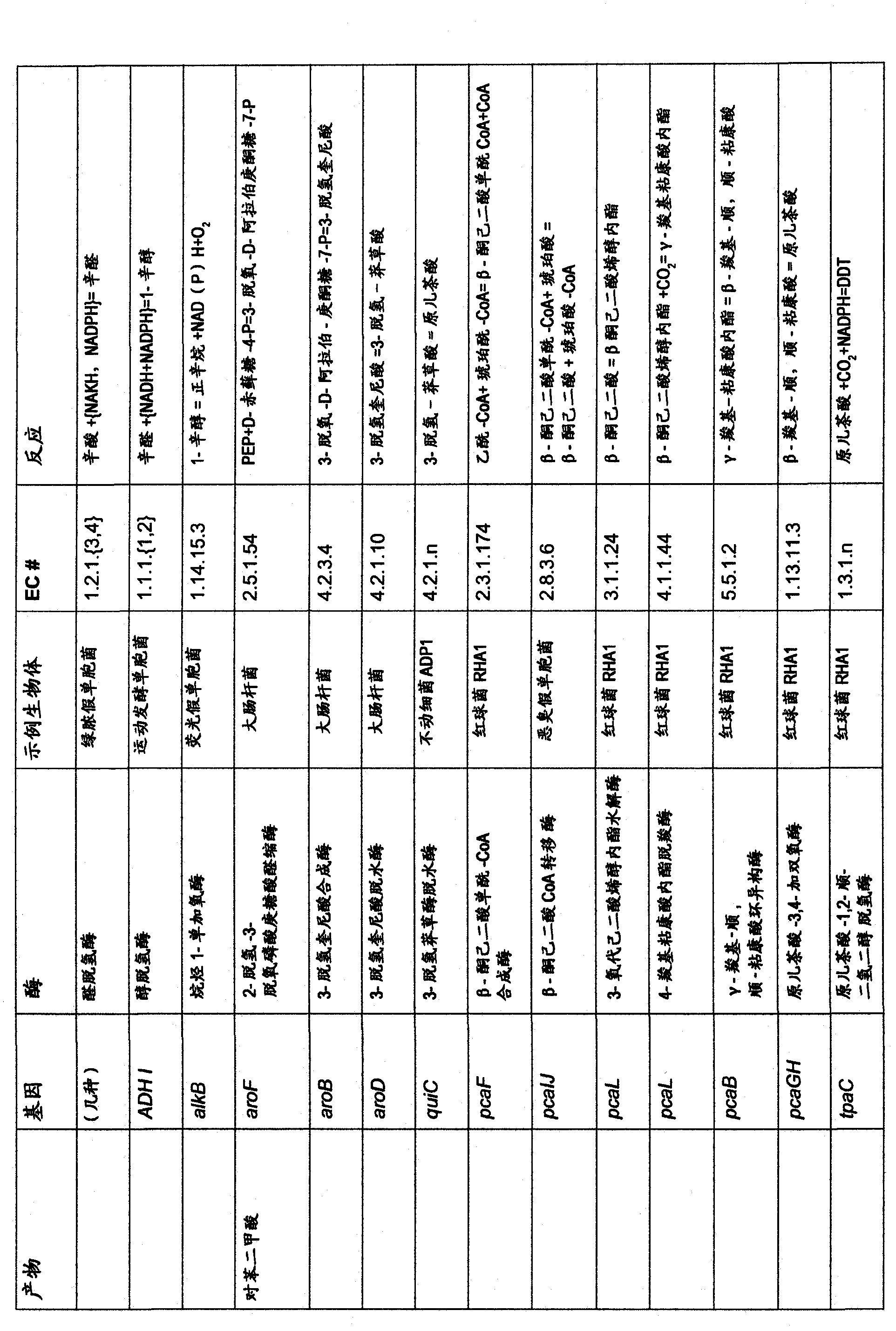
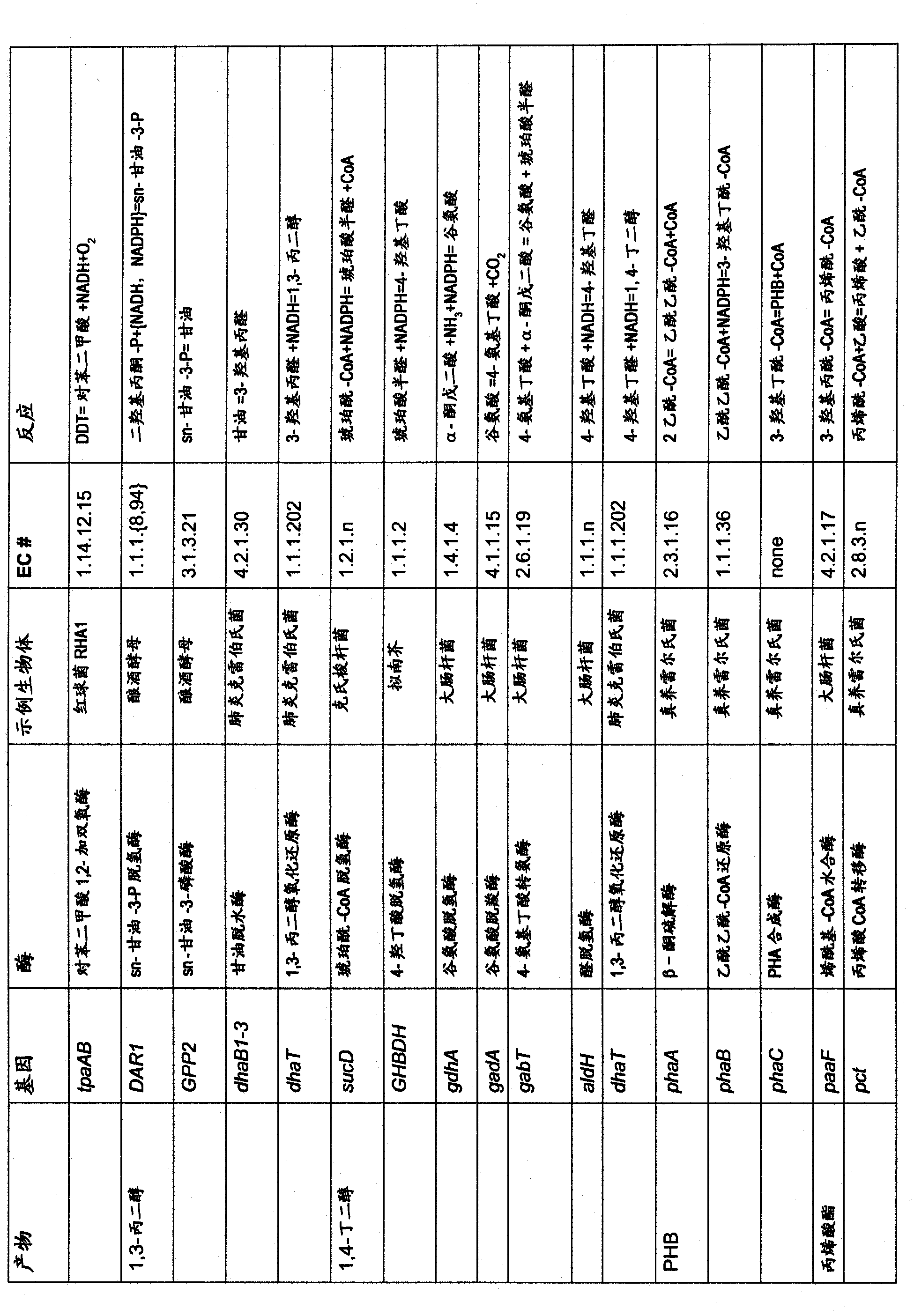
Ethanol production by microorganisms
A kind of ethanol and biological technology, which is applied in the direction of microorganisms, microorganisms, and methods based on microorganisms, and can solve problems such as inefficiency, laboriousness, and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 20
[0370] In Example 20, a method for preparing an isoprenoid compound using one or more microorganisms is described. In certain embodiments, the fuel composition prepares 3-methyl-3-buten-1-ol from 3-methyl-3-buten-1-ol by utilizing one or more microorganisms to produce 3-methyl-3-buten-1-ol. Methyl-1-butanol, and the addition of 3-methyl-1-butanol to the fuel composition produces. In other embodiments, the fuel composition prepares 3-methyl-2-buten-1-ol from 3-methyl-2-buten-1-ol by utilizing one or more microorganisms to produce 3-methyl-2-buten-1-ol. Methyl-1-butanol, and the addition of 3-methyl-1-butanol to the fuel composition produces.
[0371] Impurities
[0372] In certain aspects, the carbon-based products (e.g., ethanol, fatty acids, paraffins, isoprenoids) produced herein comprise a mixture that is typically associated with biofuels derived from triglycerides, such as fuels derived from vegetable oils and fats. than less impurities. For example, crude fatty acid ...
Embodiment 1
[0436] Plasmid Construction of Synechococcus sp. PCC 7002
[0437] Construction of pJB5: The pJB5 base plasmid was designed as an empty expression vector for recombination into Synechococcus sp. PCC7002. Two regions of homology, an upstream region of homology (UHR) and a downstream region of homology, were designed to flank the construct. These 500bp homologous regions correspond to positions 3301-3800 and 3801-4300 of UHR and DHR, respectively (Genbank accession number: NC_005025). The aadA promoter, gene sequence and terminator were designed to provide resistance to spectinomycin and streptomycin to the integrated construct. For example, pJB5 was designed with the aph2 kanamycin resistance cassette promoter and ribosome binding site (RBS). Downstream of this promoter and RBS, we designed and inserted restriction endonuclease recognition sites for NdeI and EcoRI and sites for XhoI, BamHI, SpeI and PacI. After the EcoRI site, the native terminator from the alcohol dehydroge...
Embodiment 2
[0445] Plasmid construction for Thermosynechococcus elongates BP-1
[0446] Thermosynechococcus elongates BP-1 was selected as another typical carbon dioxide fixation production host, and was modified by engineered nucleic acid to lose some genes and / or express or overexpress some genes.
[0447] Four plasmids (pJB18, pJB19, pJB20 and pJB21 ), all derivatives of pJB5, were constructed to enable homologous recombination into four different sites of the Synechococcus elongatus BP-1 genome. Specifically, the 0.5 kb upstream homology (UH) and downstream homology (DH) regions used for homologous recombination in Synechococcus PCC 7002 species in pJB5 were replaced by the following approximately 2.5 kb T. elongatus BP-1 (accession number NC_004113) Region substitutions: for pJB18, fit 831908-834231 (UH) and 834232-836607 (DH) genome; for pJB19, fit 454847-457252 (UH) and 457252-459740 (DH); for pJB20, fit 481310-483712 (UH) and 483709-486109 (DH); and for pJB21, fit 787356-789654 (...
PUM
| Property | Measurement | Unit |
|---|---|---|
| melting point | aaaaa | aaaaa |
| melting point | aaaaa | aaaaa |
| flash point | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com