Pyruvic acid producing Saccharomyces cerevisiae gene engineering bacteria and construction method and use thereof
A technology of genetically engineered bacteria and Saccharomyces cerevisiae, applied in microorganism-based methods, biochemical equipment and methods, fermentation and other directions, can solve problems such as loss of cofactors, inability to accumulate pyruvic acid, etc., achieving a simple construction method, good application prospects, Ease of use
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0022] Example 1 Producing Saccharomyces cerevisiae Engineering Bacteria of Pyruvate
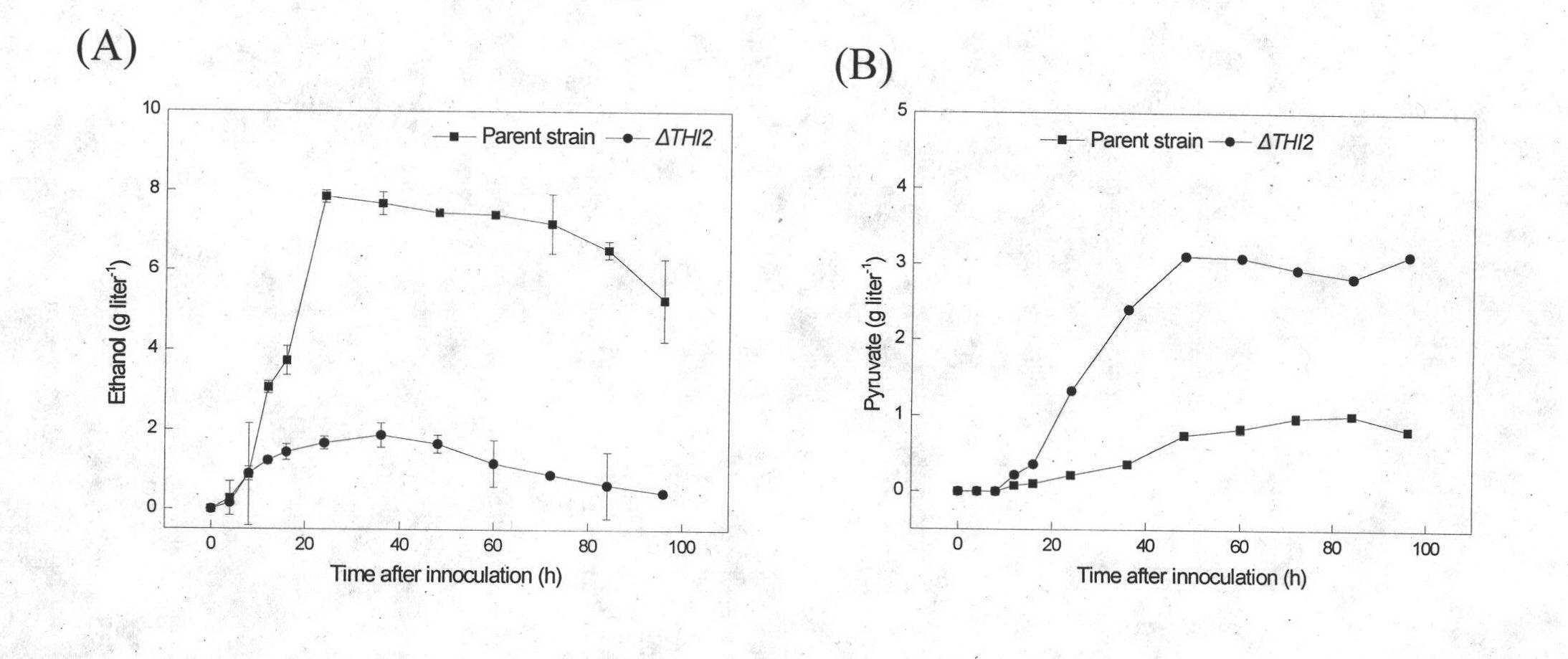
[0023] A pyruvate-producing Saccharomyces cerevisiae engineering strain, the regulatory gene THI2 in the thiamine synthesis pathway was knocked out.
Embodiment 2
[0024] Example 2 The construction method of the Saccharomyces cerevisiae engineering bacteria producing pyruvate
[0025] 1. Strains and plasmids
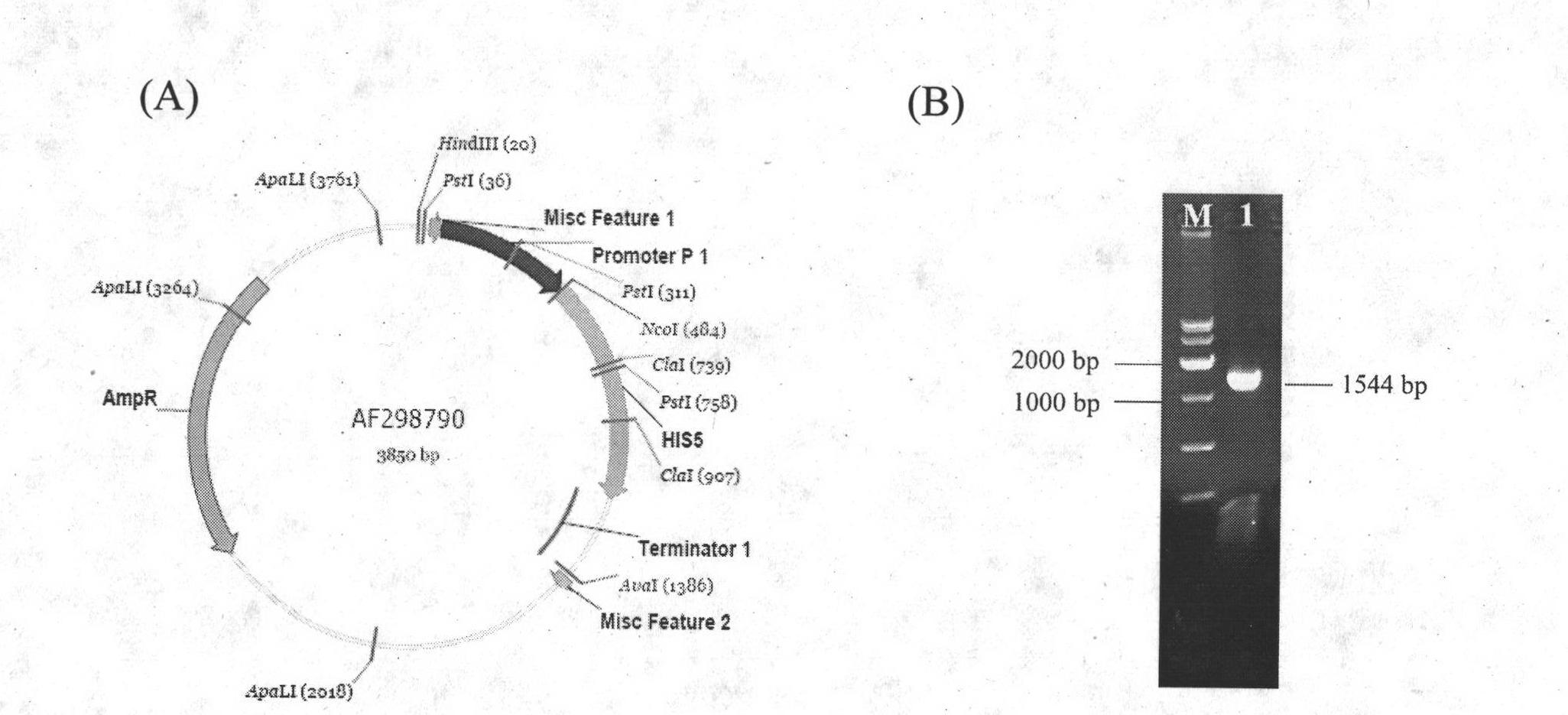
[0026] Saccharomyces cerevisiae (S.cerevisiae CEN.PK2-1C) was purchased from EUROSCARF in Europe, and its genotype was MATa ura3-52leu2-3, 112trp1-289 his3ΔMAL2-8c SUC2; plasmid pUG27 was constructed by our laboratory; plasmid pSH47 was donated by Germany.
[0027] 2. Construction of the knockout box
[0028] Plasmid pUG27 was extracted, and the plasmid was diluted 25 times as a template. The primers used to construct the knockout frame were F1:ACCACGTATATATAGCCTATATATATCCGCACTAGAACCAA-CAGCTGAAGCTTCGTACGC.R1:TGGCTTTTTTTTTCTTGAAATGAGTGAAGGGAAGGCTCAATAAGC-GCATAGGCCACTAGTGGATCTG. The obtained PCR product was purified and used to transform Saccharomyces cerevisiae.
[0029] 3. Conversion and verification
[0030] The constructed knockout frame was transformed into Saccharomyces cerevisiae (S.cerevisiae CEN.PK2-1C), verified by PCR of...
Embodiment 3
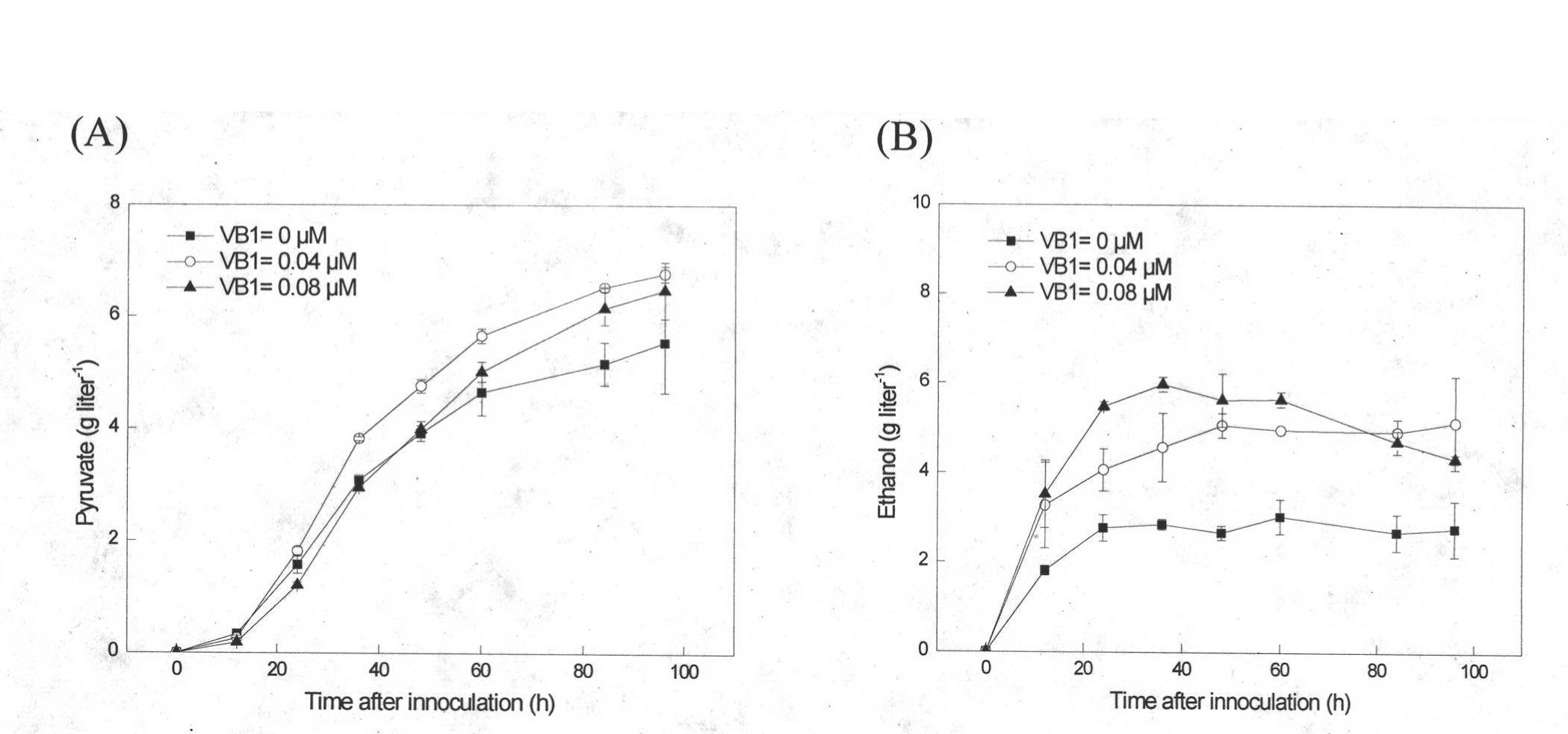
[0031] Embodiment 3 fermentation produces fumaric acid
[0032] The seeds of genetically engineered bacteria cultivated at 30°C and 200rpm for 12h were transferred to fermentation culture at an inoculum of 5%. Based on the culture at 30°C and 200rpm for 60h, different concentrations of Leu, Trp, Ura, and His were added to the medium according to the experimental requirements. . Seed culture medium: yeast extract-peptone-dextrose (YPD) medium medium, the filling volume is 20ml / 250ml. Fermentation medium (g liter-1): glucose 60, NH4Cl 7, KH2PO45, MgSO4 7H2O 0.8, trace element 10ml, pH=5.0, sterilized at 115oC for 20min, liquid volume 50ml / 250ml, CaCO340, VB1 0.04μM alone Sterilized, additionally added.
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com