A rapid detection kit for pathogenic microorganisms
A microbial and rapid technology, applied in the biological field, can solve problems such as difficulty in antibody preparation, false positive cross-reaction, and small amount of sample required
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
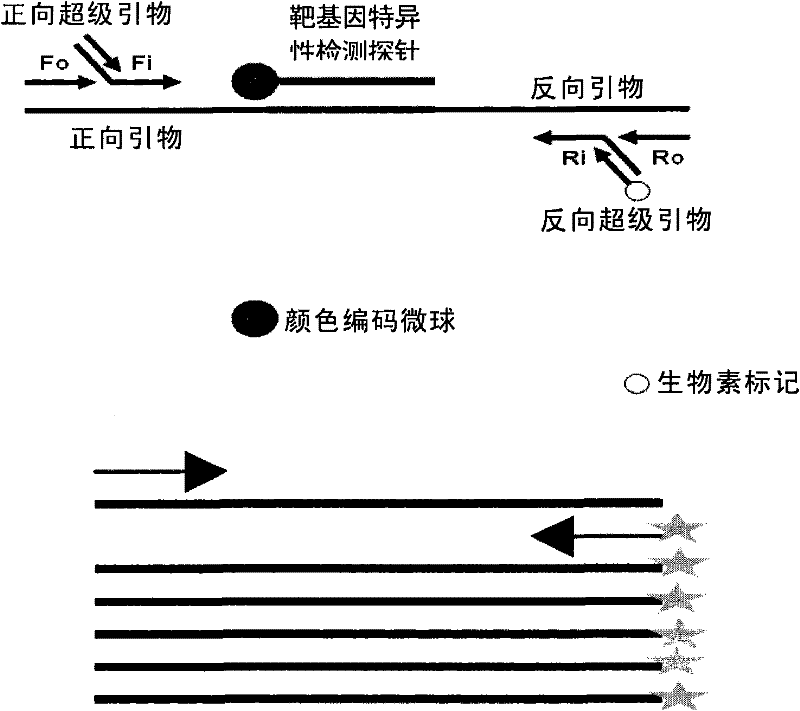
[0121] Embodiment 1, the design of primer
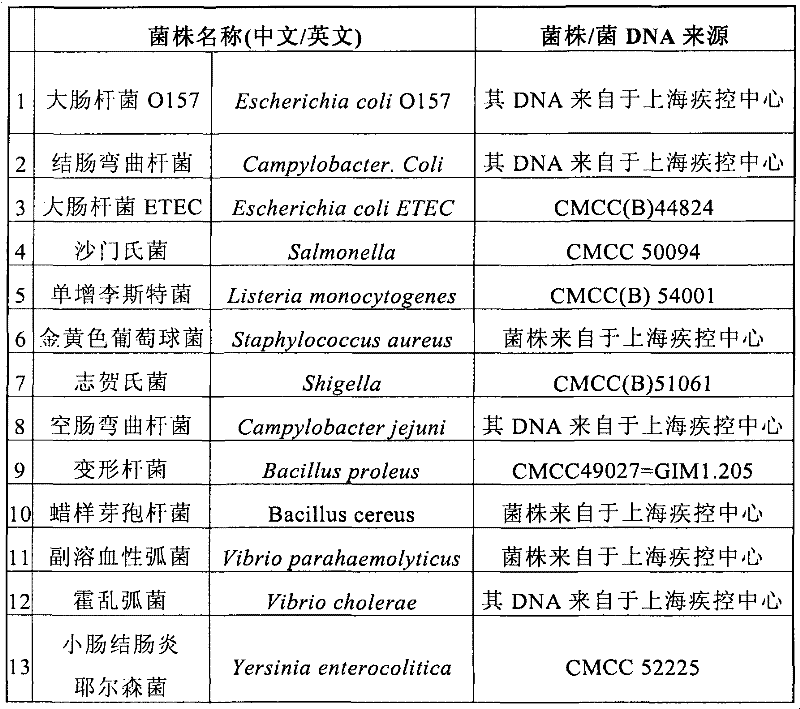
[0122] The target bacterial target gene to be detected was selected, its specific conserved sequence was determined by Blast analysis software, primers were designed by using Oligo 6 primer design software, and multiple sets of primers were designed for each detection bacterium to select the best primer combination. The selected super primers are shown in Table 2, and the selected and specific detection primer sequences for the target genes of food-borne microorganisms to be tested are shown in Table 2.
[0123] Table 2
[0124]
[0125]
Embodiment 2
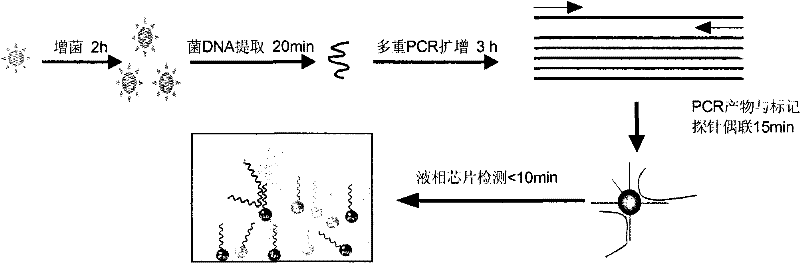
[0126] Embodiment 2, multiple PCR detection method
[0127] (1) Nucleic acid extraction
[0128] Take the bacterial solution and centrifuge at 10000rpm for 5min, collect the bacterial pellet after centrifugation, and resuspend the bacterial pellet in 100μl sterile ddH 2 In O, break the cells in a boiling water bath for 15 minutes; shake and mix well, and centrifuge at 10,000 rpm for 5 minutes; take the supernatant as a nucleic acid template for PCR.
[0129] (2)PCR
[0130] Prepare the PCR reaction solution according to the ratio in Table 3.
[0131] table 3
[0132] Reagent
µL
wxya 2 o
14
mixed primer
6
Taq Master Mix
25
nucleic acid template
5
total capacity
50
[0133] In Table 3, the mixed primers contain target gene-specific detection primers (2 pairs of primers for each target gene) and super primers, wherein relative to any target gene, the amount of the forward super primer is a...
Embodiment 3
[0137] Embodiment 3, liquid phase chip detection
[0138] The procedure for coupling probes to microspheres is as follows:
[0139] (1) Take 0.5ml of naked bead solution and centrifuge at 12,000rpm for 3 minutes;
[0140] (2) Remove the supernatant, add 50 μL of 0.1M MES solution (pH 4.5, purchased from Sigma), vortex for 10 seconds, and centrifuge at 12,000 rpm for 3 minutes;
[0141] (3) Remove the supernatant, wash once with 0.1M MES solution (pH 4.5), shake and mix, add 2.5 μL of 100 pmol / L probe solution, and vortex for 10 seconds to mix;
[0142] (4) Prepare EDC solution with 0.1M MES solution (pH4.5) to make the concentration 10mg / mL. After preparation, take 2.5μL and add it to the mixture in step 3. Vortex for 10 seconds to mix well. Stand for 30 minutes under dark conditions;
[0143] (5) 2.5 μL of freshly prepared EDC solution (purchased from Sigma) was added, vortexed for 10 seconds to mix well, and left to stand for another 30 minutes at room temperature in the ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com