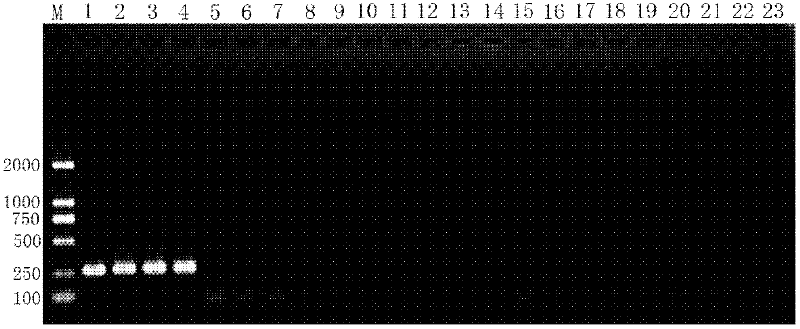Specific primer pair for assisting booklice identification and application thereof
A technology of specific primer pairs and assisted identification, which is applied in the determination/inspection of microorganisms, biochemical equipment and methods, DNA/RNA fragments, etc. and other problems, to achieve the effect of simple operation, short time-consuming, improved efficiency and accuracy
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0057] Embodiment 1, the design of specific primer pair
[0058] Based on the mitochondrial cytochrome oxidase subunit I barcode (mtDNA COI barcode) segment of ten common stored grain bookhoppers of the genus Booklice, a specific primer pair for identifying bookhoppers was designed and synthesized:
[0059] Upstream primer (sequence 1 of the sequence listing): 5'-GAACAATAATCGGGAGAGGA-3'; Tm=58.0°C, GC%=45.0;
[0060] Downstream primer (sequence 2 of the sequence listing): 5'-GTAGAAGAATTGCTGTAATAAG-3'; Tm=52.8°C, GC%=31.8.
Embodiment 2
[0061] Embodiment 2, application specific primer is paired to identify bookworm
[0062] The 22 samples were subjected to the following experiments:
[0063] 1. Extract the genomic DNA of a single booklice.
[0064] 2. Using the genomic DNA in step 1 as a template (with water as a negative control), perform PCR amplification with the specific primer pair designed in Example 1 to obtain a PCR amplification product.
[0065] PCR reaction system (26ul): 2×Taq PCR Master Mix 13ul, upstream primer (10uM) 0.5ul, downstream primer (10uM) 0.5ul, genomic DNA 2ul, ddH 2 O 10ul.
[0066] PCR reaction parameters: pre-denaturation at 95°C for 3 minutes; denaturation at 95°C for 30 sec, annealing at 50°C for 40 sec, extension at 72°C for 1 min, 32 cycles; 72°C for 8 min; storage at 4°C.
[0067] 3. Perform 1.5% agarose gel electrophoresis on 4 ul of the PCR amplified product from step 2, stain with ethidium bromide (EB), observe in the gel system, and analyze by imaging.
[0068] For th...
Embodiment 3
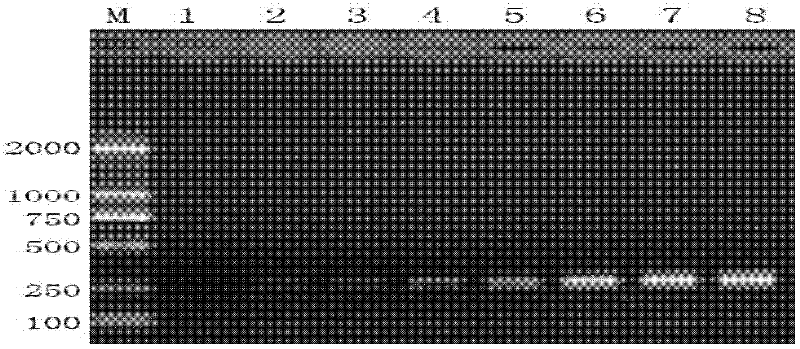
[0070] Embodiment 3, the sensitivity detection of specific primer pair
[0071] Sample 1 was subjected to the following experiments:
[0072] 1. Extract the genomic DNA of a single booklice.
[0073] 2. Detect the concentration of genomic DNA, and serially dilute it with TE buffer to obtain 8 dilutions; the concentrations of genomic DNA in the 8 dilutions are: 0.1ng / ul, 1ng / ul, 5ng / ul, 10ng / ul, 25ng / ul, 50ng / ul, 75ng / ul and 100ng / ul, dilution 1 to dilution 8 in sequence.
[0074] 3. Using each dilution as a template, perform PCR amplification with the specific primer pair designed in Example 1 to obtain PCR amplification products.
[0075] PCR reaction system (26ul): 2×Taq PCR Master Mix 13ul, upstream primer (10uM) 0.5ul, downstream primer (10uM) 0.5ul, diluent (including genomic DNA) 1ul, ddH 2 O 11ul.
[0076] PCR reaction parameters: pre-denaturation at 95°C for 3 minutes; denaturation at 95°C for 30 sec, annealing at 50°C for 40 sec, extension at 72°C for 1 min, 32 c...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com