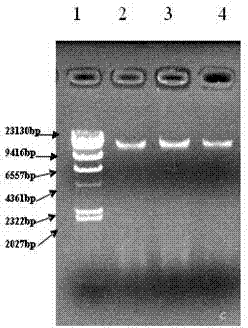
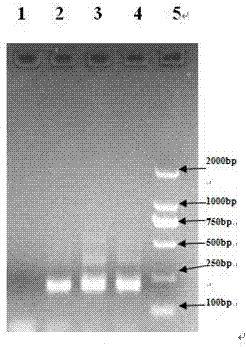
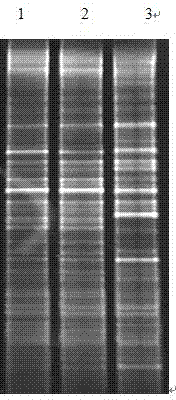
Extraction method of total genome DNA from microbes
An extraction method and genome technology, applied in the fields of environmental science and biotechnology science, can solve the problems of low DNA purity and long operation time, and achieve the effect of accurate abundance and diversity, simple operation and short time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment
[0043] Example: Microbial DNA Extraction from Shrimp Culture Pond Bottom Mud in Caojing Breeding Base, Jinshan District, Shanghai
[0044] 1. Pre-treatment of sediment samples
[0045] 1.1 Take 0.2 g of sediment sample and add 2 mL of sterilized TENP washing solution (50 mmol / L Tris, 20 mmol / L EDTA, 100 mmol / L NaCl, 1% w / v polyvinylpyrrolidone, pH = 10) Vortex to mix, bathe in 65°C water for 3 minutes, centrifuge at 8000rpm for 5 minutes, and take the precipitate. Repeat step 1 2-3 times until the supernatant becomes more transparent.
[0046] 2. Lysis of microbial cell walls in sediment samples
[0047] 2.1 Add about 6 glass beads with a diameter of 1 mm to the precipitate obtained in step 1.1, and 1 mL of lysate (1M Tris-HCl, 0.5M EDTA, 0.5M Na 3 PO4, 1.5M NaCl, 1% CTAB) and 300 μL of 50 mg / mL lysozyme solution, vortexed for 5-10 min, and placed in a water bath at 37 °C for more than 1 h.
[0048] 3. Acquisition of crude DNA from sediment samples
[0049]3.1 Add 60 ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com