Marine fish mitochondrial genome control area amplification primer, as well as design and amplification methods
A mitochondrial genome and amplification primer technology, applied in DNA preparation, recombinant DNA technology, DNA/RNA fragments, etc., can solve the problems of ineffective amplification of most marine fish and different amplification capabilities, and achieve strong amplification Abilities, effects of wide expansion range
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
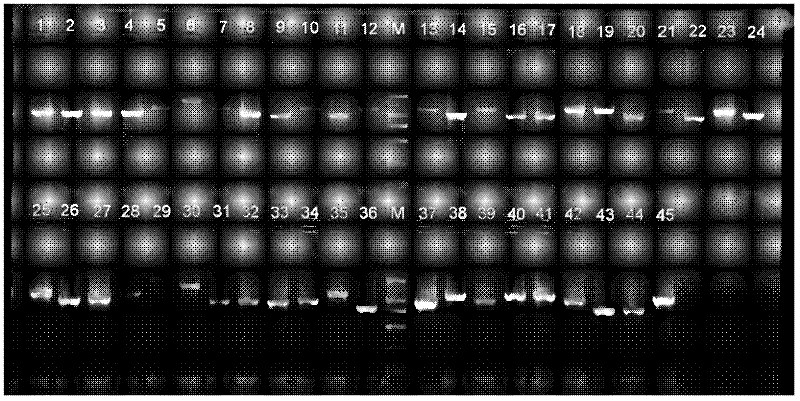
[0028] 1. Design and synthesis of primers for amplification of the mitochondrial genome control region of marine fish:
[0029] Log in to the GenBank database on the NCBI website to search for the tRNA-Thr and tRNA-Phe gene sequences of the mitochondrial genomes of marine fishes that have been determined, load the sequences into the analysis software MEGA4.1, and use the ClustalW algorithm to perform multiple sequence alignment Analyze, find the conserved sequence, load the found conserved sequence into the Premier Primer5.0 software and design the marine fish mitochondrial genome control region expansion in the manual design mode (that is, select Low under the manual option, and the specific parameters are the default settings). Booster primers: the light chain primer Marinefish-Dloop-Thr-F (SEQ ID No.1) has 20 bases: AGCACCGGTCTTGTAAACCG, located on the tRNA-Thr gene, and the heavy chain primer Marine-Dloop-Phe-R (SEQ ID No. .2) There are 21 bases: GGGCTCATCTTAACATCTTCA, loc...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com