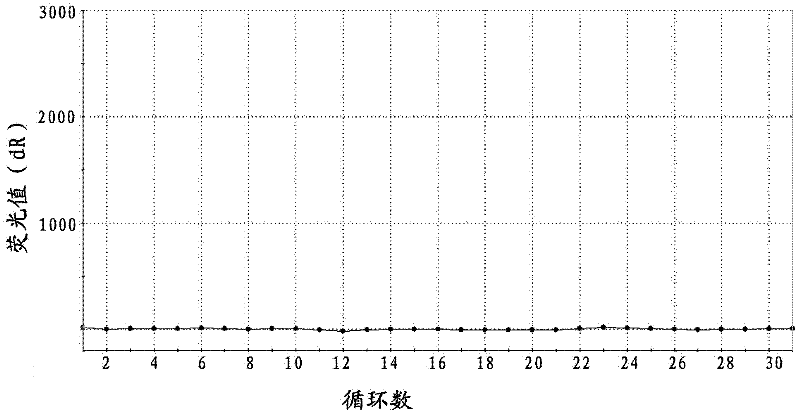
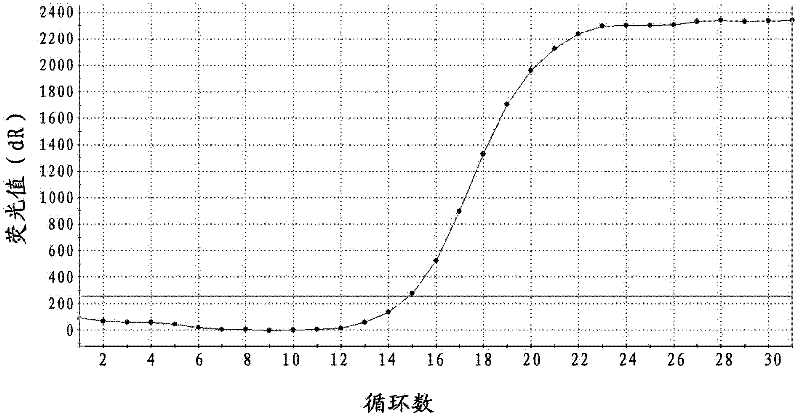
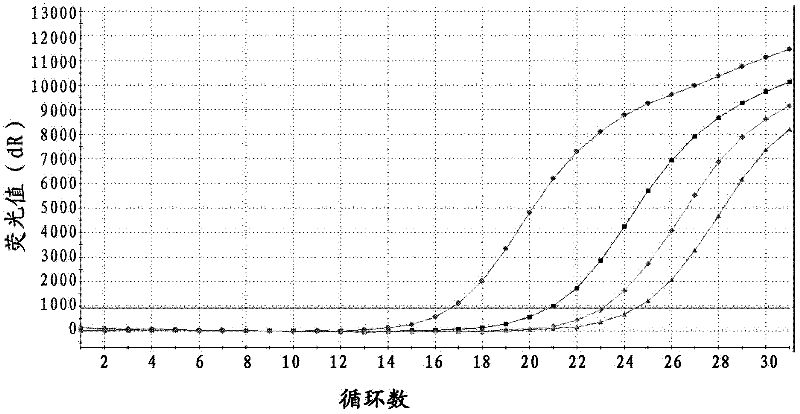
Probe, primer and kit for detecting drive mutation of PIK3CA (Phosphatidylinositol-3-kinases) gene
A detection kit and probe technology, applied in the biological field, can solve problems such as inability to achieve, and achieve the effects of high sensitivity, fast detection speed and strong specificity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0041] Take the H1047R driver mutation of exon 20 of the PIK3CA gene as an example to detect the real-time fluorescent PCR detection reaction system of the present invention. One plasmid template (including H1047R) and two cell lines were used in the experiment, namely H1650 (wild type PIK3CA gene) and H460 (wild type PIK3CA gene). The method for detecting the H1047R driver mutation of exon 20 of the PIK3CA gene by using the above fluorescent PCR is as follows:
[0042] (1) Sample DNA extraction
[0043] Cut the sample into 5-10μm, add 1mL xylene to dewax, centrifuge to collect the precipitate, add 1mL absolute ethanol to the precipitate, dry at room temperature or 37°C, add proteinase K and Buffer ATL, digest at 56°C for 1 hour, 90°C Incubate for 1h, add 200mL Buffer AL to mix well, then add 200μl absolute ethanol to mix well, carefully transfer the supernatant to a QIA 2mL spin column, centrifuge at 6000×g (8000rpm) for 1min, add 500μl Buffer AW1, 6000×g (8000rpm) ) and ce...
Embodiment 2
[0055] Using the present invention to detect clinical samples, a total of 90 cases of clinical breast cancer samples were taken and sent to our company for detection, including 70 cases of paraffin-embedded tissue samples and 20 cases of blood samples; 42 cases of males and 48 cases of females, aged 35-72 years, The average age is 58 years old. The steps for detecting PIK3CA gene driver mutations in 90 clinical samples by using the double-loop probe and fluorescent PCR system of the present invention are as follows.
[0056] (1) DNA extraction from clinical pathological samples
[0057] For fresh pathological tissue and paraffin tissue samples, cut the sample into 5-10μm, add 1mL xylene to dewax, collect the precipitate by centrifugation, add 1mL absolute ethanol to the precipitate, dry at room temperature or 37°C, add proteinase K and Buffer ATL, Digest and lyse at 56°C for 1 hour, incubate at 90°C for 1 hour, add 200mL Buffer AL and mix well, then add 200μL absolute ethanol...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com