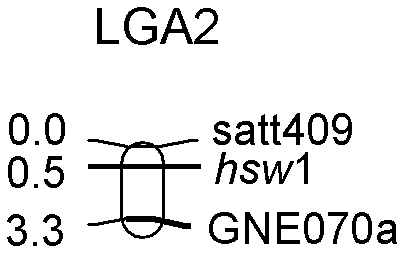Method for assisting identification of 100-seed weight related loci of soybeand special primer thereof
An auxiliary identification and 100-grain weight technology, applied in the fields of botanical equipment and methods, applications, and plant genetic improvement, can solve the problems of low selection efficiency and long breeding cycle, shorten the breeding cycle, improve the breeding efficiency and soybean yield. horizontal effect
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0031] Example 1. Acquisition of soybean 100-kernel weight-related locus (high 100-kernel weight-related locus) hsw1
[0032] 1. Primer synthesis
[0033] The sequence of the GNE070 primer pair was obtained from the SOYBASE database; the upstream forward primer sequence was: 5'-TGCTTTCCTCTATTGGTTGT-3'; the downstream reverse primer sequence was: 5'-TCTTTTCTCTGATCCATCACC-3', which was synthesized by Beijing Osaibo Biological Company.
[0034] 2. Parental genome amplification detection
[0035] The parents used to create soybean recombinant inbred line (recombinant inbred line, RIL) population were Zhonghuang 4 (National Soybean Germplasm Bank) and Zhongpin 661 (National Soybean Germplasm Bank). Zhonghuang 4 is the female parent, and its grain volume is relatively large (22.0-26.5 g / 100 grains); Zhongpin 661 is the male parent, and its grain volume is small (17.7-19.5 g / 100 grains).
[0036] Genomic DNA from parental leaves was extracted by CTAB method, and the amplification e...
Embodiment 2
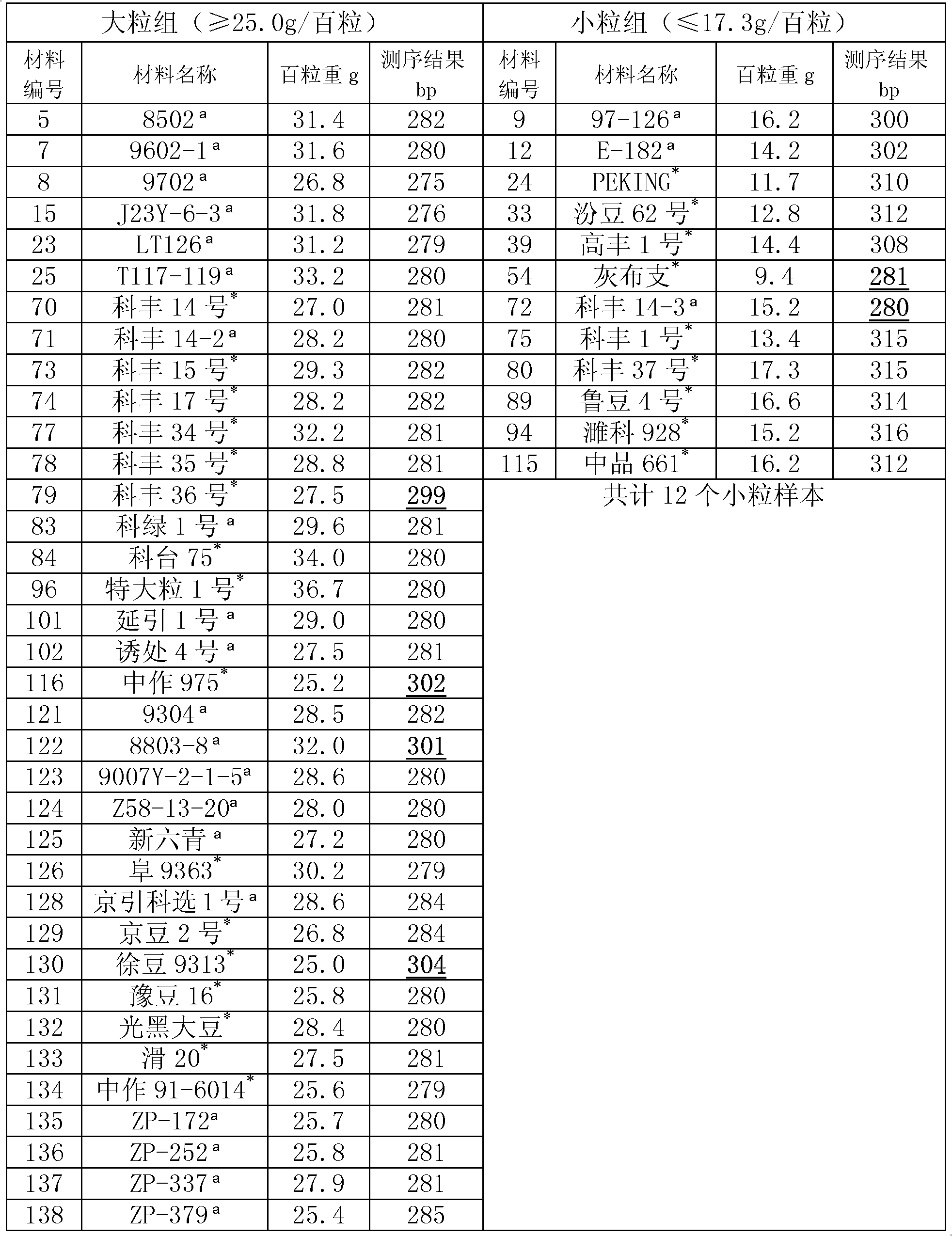
[0042] Example 2, Correlation verification between the amplification product of the GNE070a primer pair and the grain weight traits
[0043] The experimental samples were 48 soybean plant materials (homozygous varieties or lines).
[0044] The genomic DNA of each soybean plant leaf was extracted by CTAB method, and the PCR amplification experiment was carried out using the genomic DNA as a template and GNE070a primer pair. PCR reaction system (20μl): 10×Buffer 2μl (containing Mg 2+ ), dNTP (10mM) 0.4μl, upstream forward primer (5μM) and downstream reverse primer (5μM) each 2μl, genomic DNA (40ng / μl) 1μl, Taq enzyme (5U / μl) 0.5μl, the rest is ddH 2 O, add a drop of mineral oil to cover. PCR reaction program: 94°C for 5min; 35 cycles of 94°C for 60s, 53.5°C for 60s, and 72°C for 60s; 72°C for 10min.
[0045] PCR amplification products were analyzed by 9% polyacrylamide gel electrophoresis, stained with rapid silver staining and observed under ultraviolet light. The PCR ampli...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com