Quantitative biomolecule detection method
A quantitative detection method and technology of biomolecules, applied in the field of quantitative detection of biomolecules, can solve the problems of low sensitivity, cumbersome steps, and high cost, and achieve the effects of improved detection sensitivity, mild reaction conditions, and reduced detection costs
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
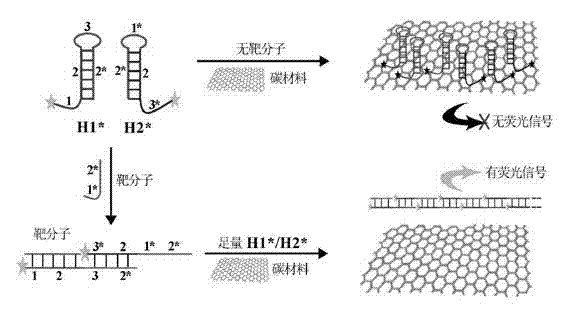
[0033] (a) Using let-7a microRNA as the target biomolecule (purchased from Dalian Bao Biological Co., Ltd.), according to its sequence (UGAGGUAGUAGGUUGUAUAGUU), design FAM fluorescently labeled H1* and H2* probes according to conventional methods, and the designed H1*, The H2* probe sequences are: H1*: AGTAGGTTGTATAGTTCAAAGTAACTATACAACCTACTACCTCA-FAM; H2*: FAM-ACTTTGAACTATACAACCTACTTGAGGTAGTAGGTTGTATAGTT.
[0034] (b) Place H1* and H2* monomer solutions with a concentration of 2 μM on a PCR instrument and heat at 95°C for 2 minutes, and then place them at room temperature in the dark for 1 hour to fully close the stem-loop structure;
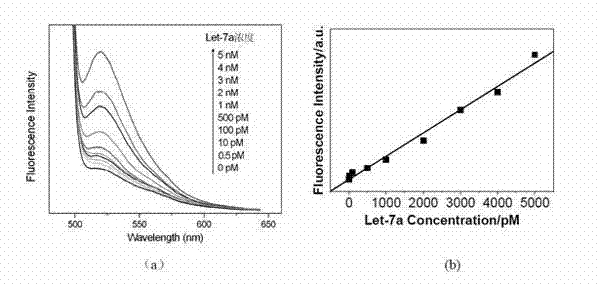
[0035](c) Take 10 RNase-free centrifuge tubes, add different concentrations of target let-7a microRNA to each tube, so that the final concentration of let-7a in each tube is 0pM, 0.5pM, 10pM, 100pM, 500pM, 1nM , 2nM, 3nM, 4nM, 5nM, and then add H1* and H2* with a final concentration of 50nM to each centrifuge tube, a certain volume of buffer sol...
Embodiment 2
[0040] (a) Let-7a mi croRNA was used as the target detection molecule, and FAM fluorescently labeled H1* and H2* probes were designed according to its sequence (UGAGGUAGUAGGUUGUAUAGUU), and the sequences were: H1*: AGTAGGTTGTATAGTTCAAAGTAACTATAC AACCTACTACCTCA-FAM; H2*: FAM -ACTTTGAACTATACAACCTACTTGAGGTAGTAGGTTGTATAGTT;
[0041] (b) Place H1* and H2* monomer solutions with a concentration of 2 μM on a PCR instrument and heat at 95°C for 2 minutes, and then place them at room temperature in the dark for 1 hour to fully close the stem-loop structure;
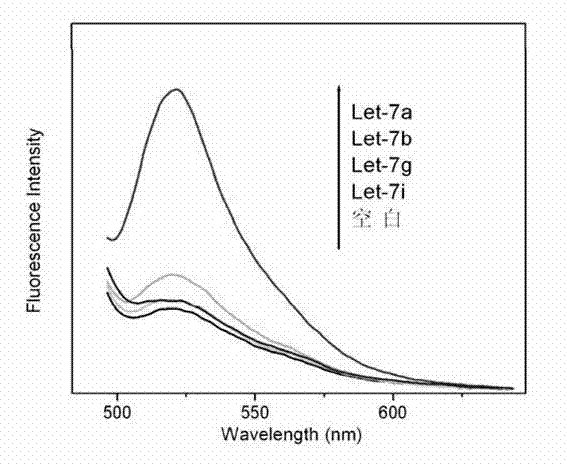
[0042] (c) Take 4 RNase-free centrifuge tubes, add let-7a, let-7b, let-7g, and let-7i at the same concentration (3nM) to each tube, and take another centrifuge tube as a blank control at the same time ; Then each centrifuge tube was added with a final concentration of 50nM H1* and H2*, a certain volume of buffer solution and water, so that the final reaction medium in each tube was 200 μL volume of SSP buffer (50mM Na 2 HPO 4 , ...
Embodiment 4
[0046] (a) Take a randomly selected sequence DNA fragment (selected from Proc.Natl.Acad.Sci.USA, 2004, 15275-15278) as the target detection molecule, and design FAM-labeled H1* and H2 according to its sequence (AGTCTAGGATTCGGCGTGGGTTAA) *Probes, the sequences are: H1*: FAM-TTAACCCACGCCGAATCCTAGACTCAAAGTAGTCTAGGATTCGGCGTG; H2*: AGTCTAGGATTCGGCGTGGGTTAACACGCCGAATCCTAGACTACTTTG-FAM;
[0047] (b) Place H1* and H2* monomer solutions with a concentration of 2 μM on a PCR instrument and heat at 95°C for 2 minutes, and then place them at room temperature in the dark for 1 hour to fully close the stem-loop structure;
[0048] (c) Take 10 centrifuge tubes, add different concentrations of target DNA molecules into each tube, so that the final concentrations are 0, 0.5pM, 10pM, 100pM, 500pM, 1nM, 2nM, 3nM, 4nM, 5nM, and then Add H1* and H2* with a final concentration of 50nM, a certain volume of buffer solution and water to each centrifuge tube, so that the final reaction medium in each t...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com