Quantitative detection method for micro RNA (Ribose Nucleic Acid)
A quantitative detection method and quality technology, applied in the determination/inspection of microorganisms, biochemical equipment and methods, fluorescence/phosphorescence, etc., can solve the problems of poor specificity and sensitivity of the three-step detection method, and achieve improved sensitivity and accuracy Effects of monitoring, enhancing sensitivity
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
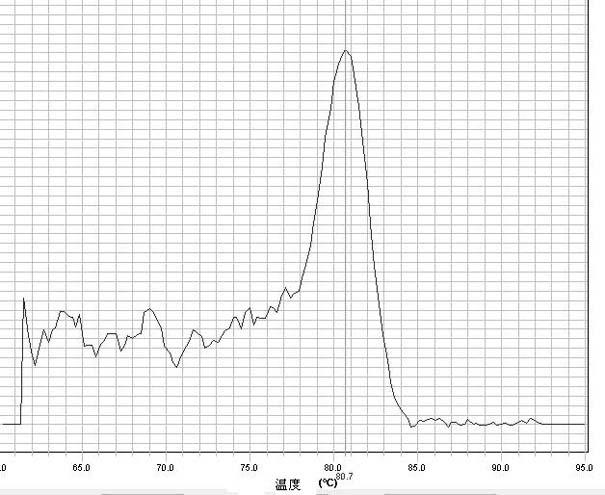
[0022] Example 1 Detection of Hsa-miR-103a content in serum
[0023] The sequence of Hsa-miR-103a is 5'-AGCAGCAUUGUACAGGGCUAUGA-3' (SEQ ID NO: 7).
[0024] 1. Extraction of total RNA from serum
[0025] Take 2ml of blood from each of two adults, centrifuge after coagulation, and finally take 1ml of the upper serum and place it in a 1.5ml RNase / DNase-free centrifuge tube.
[0026] Use the RNA extraction kit (Beijing Kuangbo Biotechnology Co., Ltd.) to extract total RNA from serum, and add 1 μl (20 nM) of the sequence 5'-CAACCTCCTAGAAAGA-3' (SEQ ID NO: 1) to each 250 μl of serum. 1 (synthesized by Shanghai Sangon Bioengineering Technology Co., Ltd.) to monitor the quality of RNA extraction from serum. The concentration of extracted total RNA was determined using Thermo NanoDrop 2000c.
[0027] 2. Three-step method for quantitative detection of HSA-miR-103a in serum
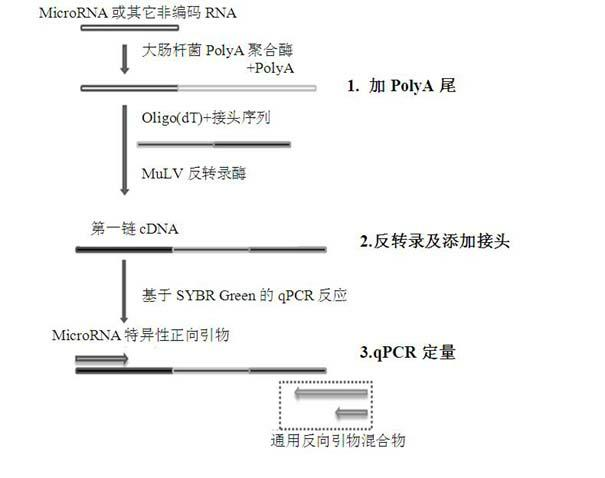
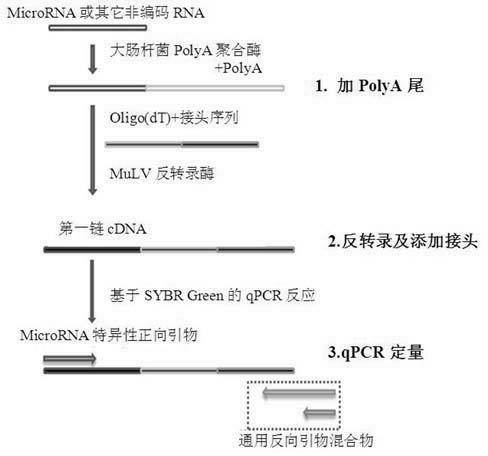
[0028] (1) Add polyA tail:
[0029] i. Prepare the polyA-tailed reaction solution in an RNase-free PCR tube ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com