A method for detecting active demethylation of DNA
A demethylation and methylation technology, applied in recombinant DNA technology, biochemical equipment and methods, DNA/RNA fragments, etc.
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
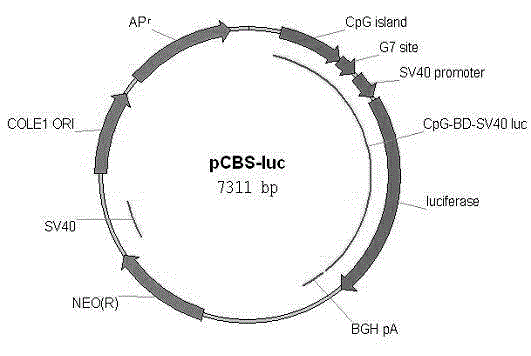
[0036] Example 1. Construction and in vitro methylation of artificial CpG island reporter gene carrier PCBS-luc
[0037] 1. Design of PCBS-luc
[0038] Using the method of Escherichia coli recombinant plasmid construction, with PCDNA3.1 as the backbone, luciferase reporter gene, SV40 promoter, BD domain binding site, and a high CpG fragment derived from Streptomyces coelicolor were inserted in sequence to obtain (named PCBS -luc).
[0039] 2. In vitro methylation of PCBS-luc
[0040] PCBS-luc with restriction endonuclease Pst I was linearized by single enzyme digestion and incubated at 37 degrees for 2 hours. Afterwards, the product was purified (using the Axygen Gel Recovery Kit), half of the recovered product was used for methylation treatment, and the other half was not treated as a non-methylated control. In vitro methylation treatment using M.Sss I (NEB Corporation). After the methylation treatment, the product was purified again in the same way as before. rest...
Embodiment 2
[0043] Example 2: Screening of monoclonal stably transfected cell lines
[0044] 1. Determine the optimal drug concentration for screening
[0045] The optimal drug concentration refers to the lowest concentration that can cause all cells to die. The selected drug is hygromycin B (Hygromycin B), its concentration is 50mg / ml. The cells used were HEK293. Specific steps are as follows:
[0046] (1) Resuscitate HEK293 cells into a 6cm dish, passage when they are full, transfer 3ml of 18-20ul per well into a 96-well plate, and make up 100ul / well.
[0047] (2) According to the literature, the optimal concentration of this drug for HEK293 cells is 200ug / ml, and it is also recommended to be 100ug / ml. Therefore, the pre-experimental drug concentrations were selected as 80 ug / ml, 100 ug / ml, 150 ug / ml, and 200 ug / ml to determine the final suitable drug concentration for this experiment.
[0048] (3) After the cells in the 96-well plate are well-passed, pressurize and screen. After 6...
Embodiment 3
[0069] Example 3: Identification of methylated monoclonal stably transfected cell lines
[0070] 1. Identification of Luciferase Activity in Methylated and Unmethylated Monoclonal Stably Transfected Cell Lines
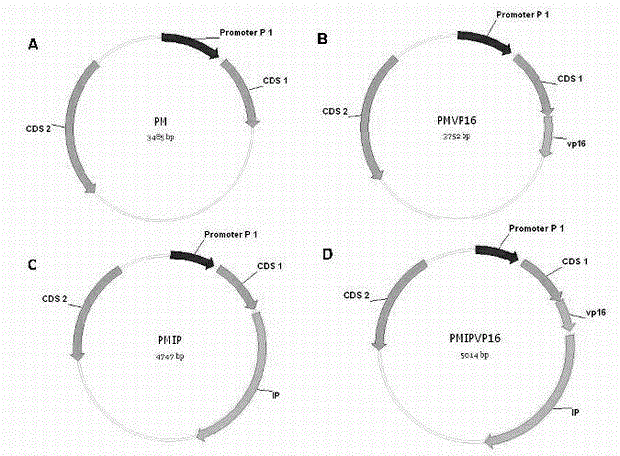
[0071] (1) Transfer the screened stably transfected monoclonal cell lines into 96-well plates, and transfect within 12-16 hours. PM is used as the empty control.
[0072] (2) After 72 hours, the luciferase value was measured and calculated.
[0073] (3) The result is as follows Figure 4 Shown in C and D, C is the transfection response luciferase activity of methylated monoclonal cells to PMVP16, D is the transfection response luciferase activity of non-methylated monoclonal cells to PMVP16 ; Select methylated cell clone 7# with higher luciferase activity, methylated cell clone 9# with lower luciferase activity, and unmethylated cell 8# for further identification.
[0074] 2. Identify the degree of methylation in methylated cell lines and non-methylated cell lines...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com