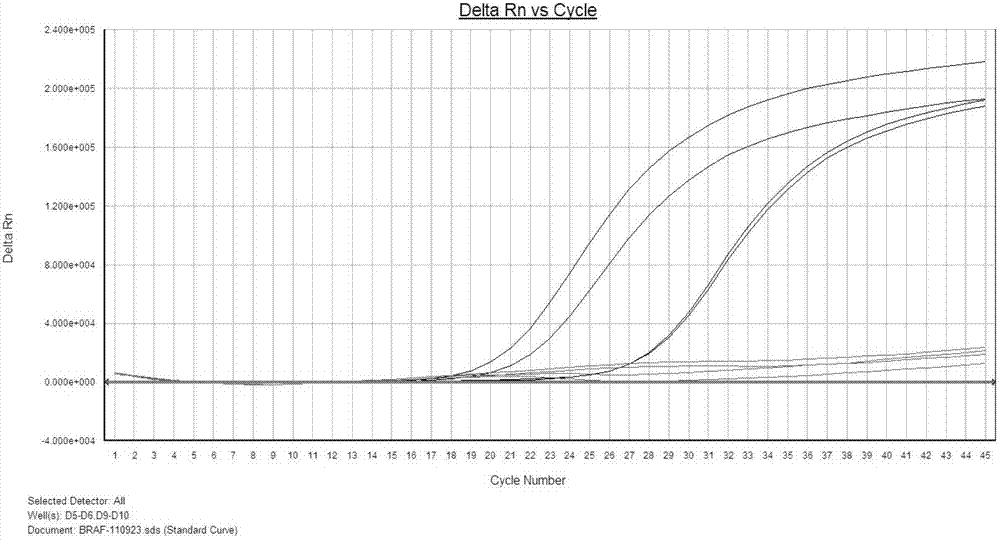
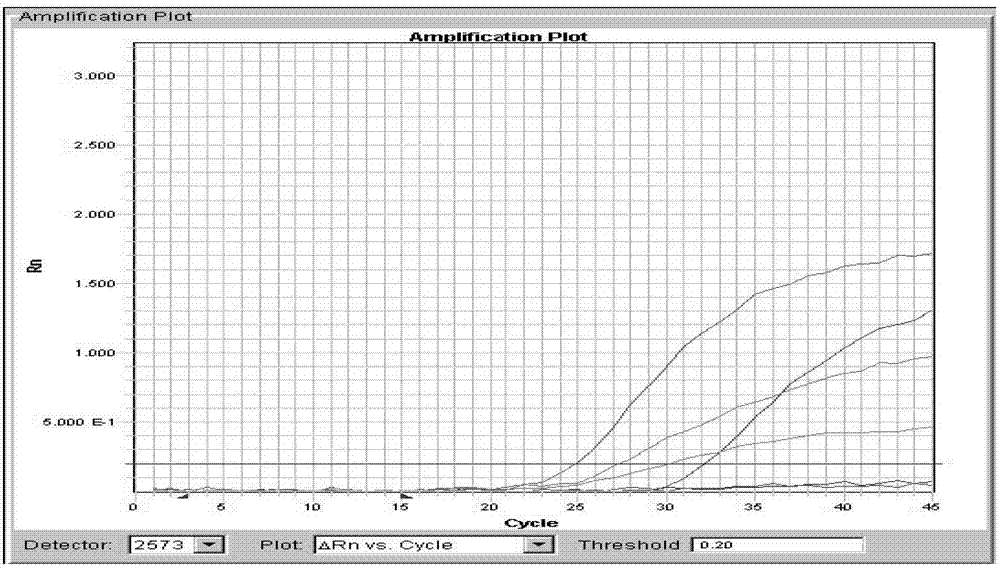
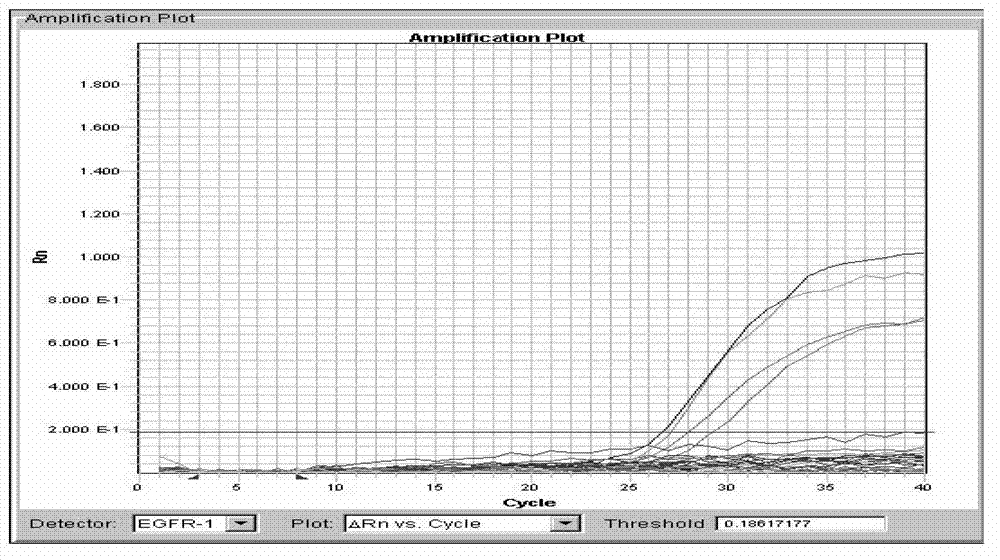
PIK3CA gene mutation fluorescence quantitative PCR genotype detection kit and detection method
A detection kit and fluorescence quantitative technology, applied in the field of molecular biology, can solve the problems of complex operation process, insufficient detection sensitivity and time-consuming
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0070] Example 1: Detection of base substitution mutations at positions 542 (G→A mutation) and 545 (G→A mutation) of exon 9 of PIK3CA gene
[0071] Sample Description: Collect tumor tissues from 50 patients with clinical pathological diagnosis of intestinal cancer (adenocarcinoma), all of whom have not received radiotherapy and chemotherapy before operation. Genomic DNA is extracted from the tumor tissues of case specimens for the following experimental applications. Prior to this, the tumor tissue content of the test specimen must be evaluated first, and the cases with tumor cell components greater than 30% are selected for the implementation test of the kit of the present invention.
[0072] Design a fluorescent probe and a pair of common primers that can specifically detect base substitution mutations at the 542nd (G→A mutation) and 545th positions (G→A mutation) of exon 9 of the PIK3CA gene, and one pair of common primers, plus one for simultaneous coverage The wild-type f...
Embodiment 2
[0085] Example 2: Detection of base substitution mutation at position 1047 (A→G) of exon 20 of PIK3CA gene
[0086] Design a fluorescent probe and a pair of common primers that can specifically detect the base substitution mutation at the 1047th (A→G) site of exon 20 of the PIK3CA gene, plus a wild-type fluorescent probe sequence to detect this site.
[0087] Among them, the PCR primers are:
[0088] Exon20F: 5'-TAC ATT CGA AAG ACC CTA GCC T-3' (SEQ ID NO: 6);
[0089] Exon20R: 5'-ATC CAT TTT TGT TGT CCA GCC-3' (SEQ ID NO: 7)
[0090] Fluorescent probes are:
[0091] Exon20-1047MT: 5'-FAM-TGA TGC ACG TCA TGG T-BHQ 1-3' (SEQ ID NO:8);
[0092] Exon20-1047WT: 5'-HEX-TGA TGC ACG TCA TGG T-BHQ1-3' (SEQ ID NO: 9).
[0093] Wherein, SEQ ID NO: B3 is a probe for detecting the base corresponding to the 1047 site of the 20th exon of the mutant PIK3CA gene, and SEQ ID NO: B4 is a wild-type fluorescent probe covering the 1047 site of the 20th exon of the PIK3CA gene. The needle sequ...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com