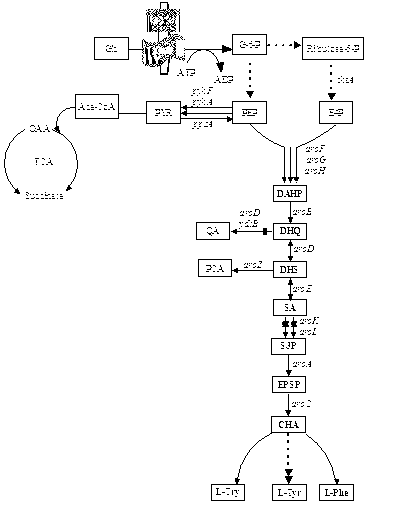
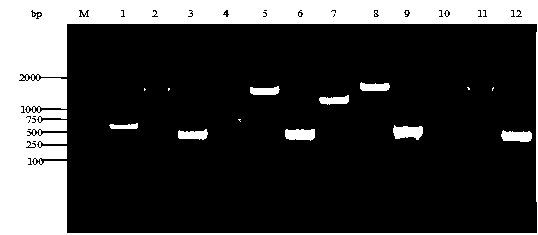
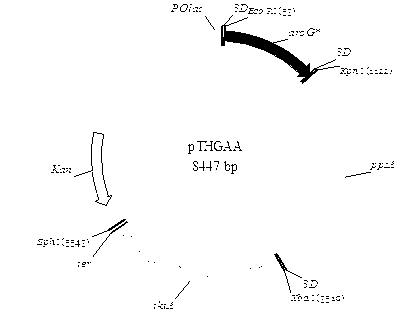
Escherichia coli recombinant strain producing shikimic acid, and construction method and application thereof
A technology of Escherichia coli and construction methods, applied in the field of microbial genetic engineering, to achieve the effect of high-efficiency accumulation
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0036] Example 1, construction of mutant strain CICIM B0013 SA4
[0037] 1.1 Mutant strain CICIM B0013·SA1 ( △aroL ) build
[0038] used to aroL Primers designed upstream and downstream of the gene aL up, aL dn amplifies the chromosomal DNA of Escherichia coli aroL gene, the fragment size is 575 bp. The PCR amplification system is 50 μL (Taq enzyme 1 μL, Taq enzyme buffer 5 μL, dNTPs 4 μL, upstream and downstream primers 1 μL, template 1 μL, ddH 2 O 17μL), the conditions are: 94°C, 5min; 94°C, 30s; 52°C, 1min; 72°C, 1min, 30 cycles. The PCR product was cloned into the pMD18-T-simple vector to obtain the recombinant plasmid pMD- aroL . Using the recombinant plasmid as a template, primers L verup, L verdn for inverse PCR amplification. The PCR amplification system was 50 μL, and the conditions were: 94°C, 5min; 94°C, 30s; 58°C, 1min; 72°C, 3.5min, 30 cycles. PCR product with difGm The fragments were ligated to obtain the recombinant plasmid pMD- aroL ':: ...
Embodiment 2
[0070] Embodiment 2, construction of recombinant plasmid pTHGAA
[0071] 2.1 Cloning of recombinant plasmid pTHG
[0072] Using the genome of Escherichia coli CICIM B0013 as a template, primers aoGm-up, aoGm-dn, aoG-up, aoG-dn were used to perform overlapping PCR to amplify the site-directed mutation aroG* gene. The primer sequences are as follows:
[0073] aoGm-up: 5′-gtgagtttctcaatatgatcaccc-3′
[0074] aoGm-dn: 5′-tggggtgatcatattgagaaact-3′
[0075] aoG-up: 5′-ccggaattcaggaggccatccatgaattatcagaacgac-3′
[0076] aoG-dn: 5′-cggggtaccttacccgcgacgcgcttttact-3′
[0077] Fragment P1 was obtained by PCR amplification with primers aoG-up and aoGm-dn, and fragment P2 was obtained by second-round PCR amplification with primers aoGm-up and aoG-dn. The PCR amplification system is 50 μL (Taq enzyme 1 μL, Taq enzyme buffer 5 μL, dNTPs 4 μL, upstream and downstream primers 1 μL, template 1 μL, ddH 2 O 17 μL), the PCR amplification conditions were: 94°C, 5min; 94°C, 30s; 56°C, 1min; ...
Embodiment 3
[0091] Embodiment 3, the acquisition of shikimic acid expression strain
[0092] The mutant strain CICIM B0013 SA4 ( △aroL , △aroK , △ptsG , △ydiB ) as a host bacterium, the recombinant expression plasmid pTHGAA obtained in Example 2 was transformed into it by electric shock transformation method, and the Escherichia coli host bacterium SA4 / pTHGAA expressing shikimic acid was obtained.
PUM
| Property | Measurement | Unit |
|---|---|---|
| solubility (mass) | aaaaa | aaaaa |
Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com