Method for establishing and identifying core fucosyltransferase gene silencing cell model
A gene silencing and cell model technology, applied in the field of biological function research, can solve problems such as no reports
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
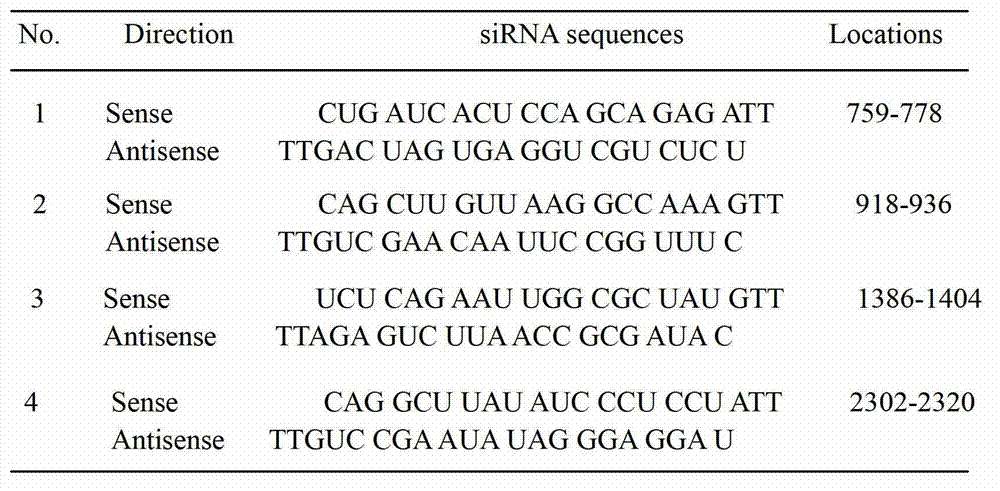
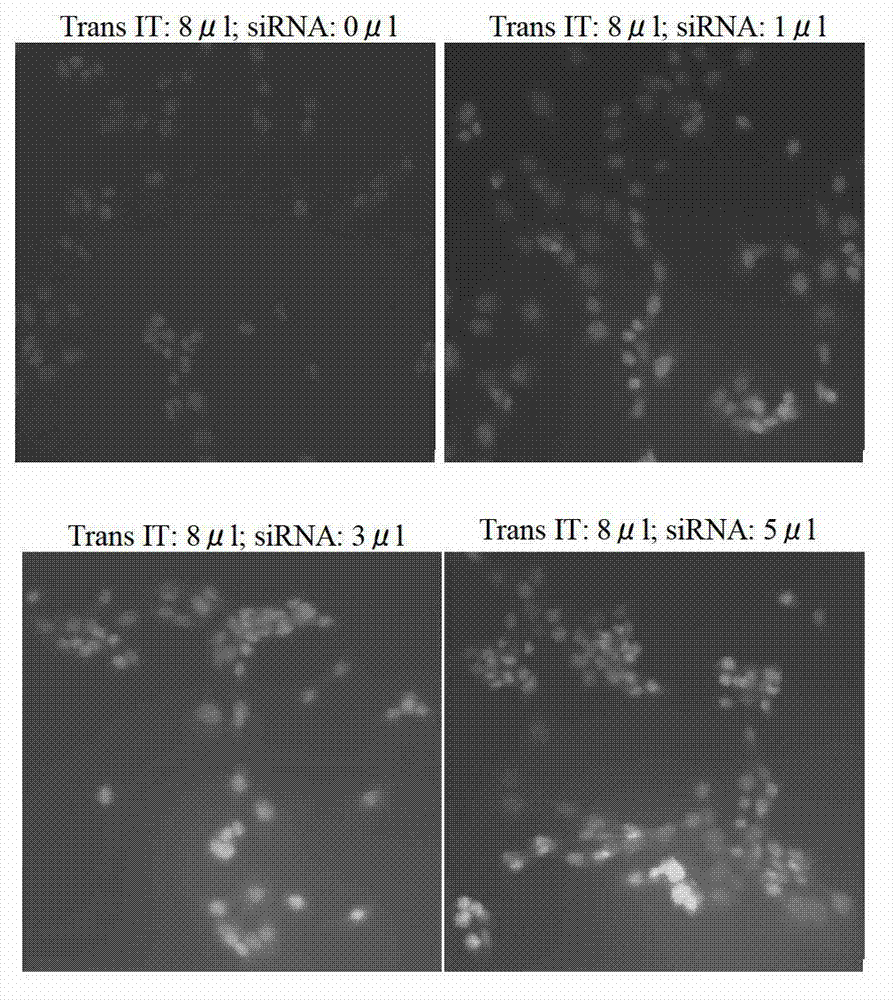
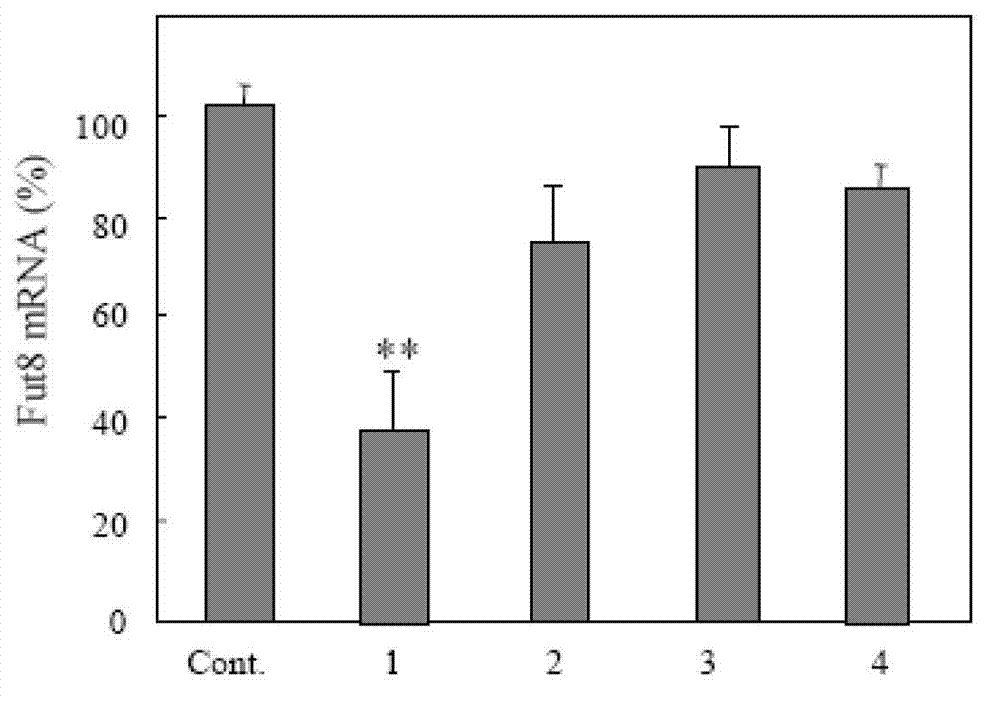
[0044] (1) Selection of Fut8 siRNA fragments Four 21-nucleotide double-stranded siRNA sequences (19 nucleotides + overhang TT) were designed using the online http: / / rnaidesigner.invitrogen.com / sirna software program. Select 4 kinds of siRNA sequences respectively from Fut8 gene (GenBank Accession NM_016893) ( figure 1 ). Using TransIT-TKO transfection reagent (Mirus Co. Madison, USA) to transfect 4 double-stranded siRNAs into the target cells, after 24 hours, screen the most effective siRNA sequences that specifically inhibit the expression of Fut8. The ratio of TransIT-TKO transfection reagent to double-stranded siRNA is 8:3 ( figure 2 ). After measuring the enzyme activity, Fut8siRNA (759-778:CUG AUC ACU CCA GCA GAGATT) was finally determined to be the best sequence ( image 3 ).
[0045] (2) Construction of recombinant retroviral vector containing Fut8 gene The construction of pSINsi-hU6-Fut8 was designed into a double-stranded hairpin structure with siRNA hairpin Oli...
Embodiment 2
[0051] (1) Selection of Fut8 siRNA fragments Four 21-nucleotide double-stranded siRNA sequences (19 nucleotides + overhang TT) were designed using the online http: / / rnaidesigner.invitrogen.com / sirna software program. Select 4 kinds of siRNA sequences respectively from Fut8 gene (GenBankAccession NM_016893) ( figure 1 ). Using TransIT-TKO transfection reagent (Mirus Co. Madison, USA) to transfect 4 double-stranded siRNAs into the target cells, after 24 hours, screen the most effective siRNA sequences that specifically inhibit the expression of Fut8. The ratio of TransIT-TKO transfection reagent to double-stranded siRNA is 8:5 ( figure 2 ). After measuring the enzyme activity, Fut8siRNA (759-778:CUG AUC ACU CCA GCA GAGATT) was finally determined to be the best sequence ( image 3 ).
[0052] (2) Construction of recombinant retroviral vector containing Fut8 gene The construction of pSINsi-hU6-Fut8 was designed into a double-stranded hairpin structure with siRNA hairpin Olig...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D
- Intellectual Property
- Life Sciences
- Materials
- Tech Scout
- Unparalleled Data Quality
- Higher Quality Content
- 60% Fewer Hallucinations
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2025 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com