Crystal structure of enterovirus 71 type 2A protease and application of crystal structure in drug design
A technology of enterovirus and protease, applied in the field of crystal structure of protease, can solve the problems of complex transmission route, no enterovirus 71 drug, strong infectivity, etc., and achieve far-reaching effects
- Summary
- Abstract
- Description
- Claims
- Application Information
AI Technical Summary
Problems solved by technology
Method used
Image
Examples
Embodiment 1
[0037] Example 1 Expression and purification of enterovirus 71 type 2A protease
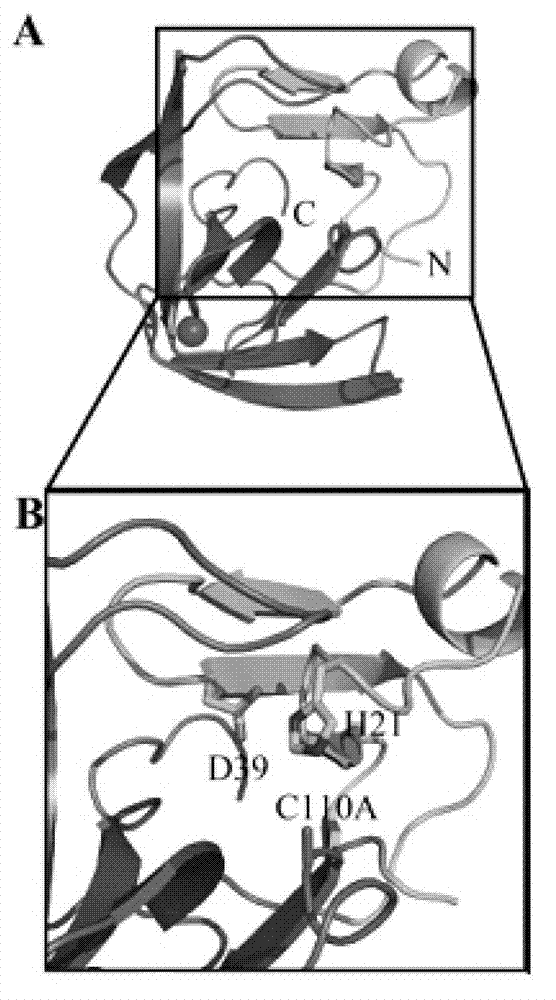
[0038]Based on the E2004104-TW-CDC virus strain of enterovirus 71 (EV71), the DNA sequence encoding EV712A protease was artificially synthesized. A single point mutation was performed on the DNA sequence by PCR experiment, and the cysteine (Cys) at the 110th position was mutated to alanine (Ala), as shown in SEQ ID NO.2. Afterwards, the mutated DNA sequence was inserted into the pGEX-4T-1 vector to construct a recombinant plasmid capable of expressing 2A protease with a nitrogen-terminal glutathione S-transferase (GST) tag.
[0039] Transform the recombinant plasmid into Escherichia coli pLysS competent cells, pick the obtained transformant clones and transfer them to appropriate amount of LB medium (adding chloramphenicol and ampicillin to the final concentration of 50 μg / ml respectively), culture at 37°C When the absorbance at 600 nm is 0.6, add isopropyl-β-D-thiogalactopyranoside (IPTG) ind...
Embodiment 2
[0040] Example 2 Crystallization and structural analysis of enterovirus 71 type 2A protease
[0041] 2.1 Crystallization conditions
[0042] Crystallization was carried out by the hanging drop method. The volume ratio of the protein solution in the crystallization droplet to the crystallization reagent was 1:1, and the protein concentration in the protein solution was 10mg / ml. Crystals grew after three days. The crystallization condition was 0.1M hydroxyethylpiperazine B Sulfuric acid (HEPES) pH 7.5, 20% isopropanol, 10% polyethylene glycol 4000 (PEG4000), crystals were stored at 16°C.
[0043] In order to obtain the complex formed by the 2A protease and the small molecular compound, the protease and the small molecular compound were mixed in an ice bath at a molar ratio of 1:5 for 2 hours. The co-crystallization conditions of protease and small molecule compound are the same as when no compound is added.
[0044] 2.2 Data collection and structure analysis
[0045] Data col...
Embodiment 3
[0049] The substrate binding groove of embodiment 3 enterovirus 71 type 2A protease
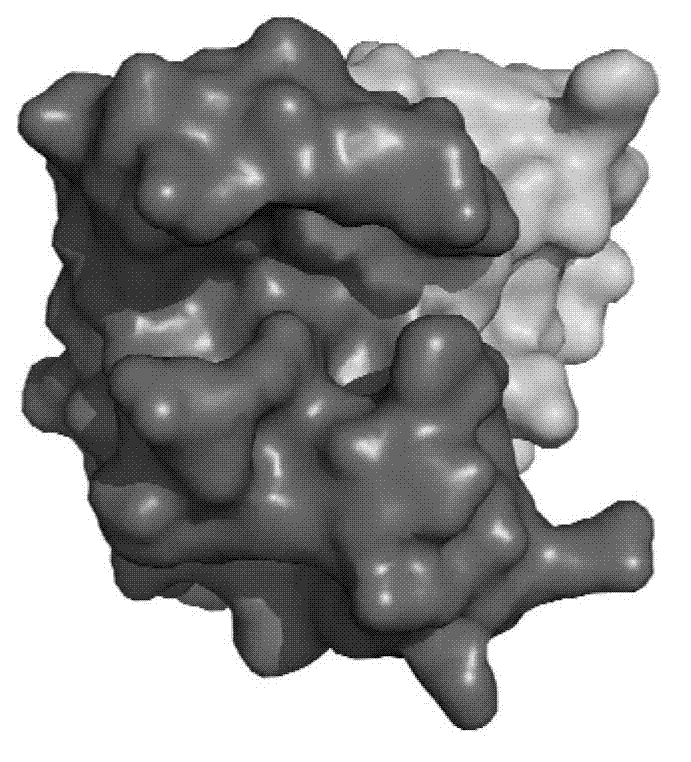
[0050] 2A The nomenclature of the protease active site: take the cleaved peptide bond on the substrate polypeptide as the reference point, and the amino acids to the left are named P1, P2, P3, ... P N , named P1', P2', P3', ...P in sequence to the right N ’; The part of the protease that accommodates these polypeptide amino acids is correspondingly named as the site S1, S2, S3...S N and S1', S2', S3'...S N '.
[0051] The substrate-binding groove of enterovirus type 2A protease is long enough to accommodate P1-P5 amino acid residues. If the catalytic triad is placed on the right, the lower end of the substrate-binding groove is formed by a carbon-terminal domain with an active residue, Cys110, located below the edge of the narrow region of the substrate-binding groove . The nitrogen-terminal domain with active residues His21 and Asp39 is at the upper right of the substrate-binding groove...
PUM
 Login to View More
Login to View More Abstract
Description
Claims
Application Information
 Login to View More
Login to View More - R&D Engineer
- R&D Manager
- IP Professional
- Industry Leading Data Capabilities
- Powerful AI technology
- Patent DNA Extraction
Browse by: Latest US Patents, China's latest patents, Technical Efficacy Thesaurus, Application Domain, Technology Topic, Popular Technical Reports.
© 2024 PatSnap. All rights reserved.Legal|Privacy policy|Modern Slavery Act Transparency Statement|Sitemap|About US| Contact US: help@patsnap.com